Filter results
Category
- (-) Earth System Science (136)
- (-) Visual Analytics (6)
- (-) Electric Grid Modernization (2)
- Scientific Discovery (307)
- Biology (198)
- Human Health (102)
- Integrative Omics (73)
- Microbiome Science (42)
- Computational Research (23)
- National Security (21)
- Computing & Analytics (14)
- Chemistry (10)
- Energy Resiliency (9)
- Data Analytics & Machine Learning (8)
- Materials Science (7)
- Chemical & Biological Signatures Science (5)
- Computational Mathematics & Statistics (5)
- Weapons of Mass Effect (5)
- Atmospheric Science (4)
- Coastal Science (4)
- Ecosystem Science (4)
- Renewable Energy (4)
- Data Analytics & Machine Learning (3)
- Plant Science (3)
- Cybersecurity (2)
- Distribution (2)
- Energy Efficiency (2)
- Energy Storage (2)
- Grid Cybersecurity (2)
- Solar Energy (2)
- Bioenergy Technologies (1)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Omics (21)
- Soil Microbiology (21)
- sequencing (13)
- Genomics (10)
- Metagenomics (9)
- Microbiome (8)
- Fungi (6)
- High Throughput Sequencing (6)
- Imaging (6)
- Mass Spectrometry (6)
- Mass Spectrometer (5)
- metagenomics (4)
- Microscopy (4)
- Sequencer System (4)
- Sequencing (4)
- soil microbiology (4)
- Spectroscopy (4)
- Climate Change (3)
- IAREC (3)
- metabolomics (3)
- PerCon SFA (3)
- Proteomics (3)
- Synthetic Biology (3)
- Viruses (3)
- Cybersecurity (2)
- Electrical energy (2)
- Lipidomics (2)
- microbiome stability (2)
- omics (2)
- species volatility (2)
Category
Category
Category
Category
Category
Category
Category
Category
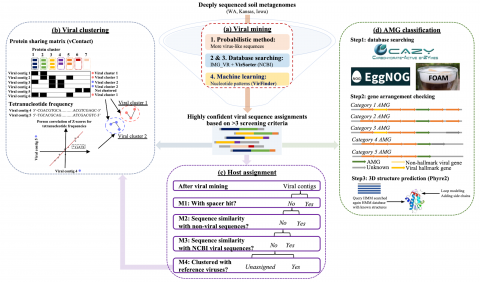
Viral genome assembly annotations from terabase metagenomes (TmG.1.0) from uncultivated soil collections from the IAREC field sites in Washington (IAREC), Kansas (KBPS), and Iowa (COBS), USA. Uncultivated virus Genomes (identified from soil metagenome datasets) Data DOI Package.
Category
Code pertaining to the Soil Microbiome SFA Project publication data visualizations 'DNA viral diversity, abundance and functional potential vary across grassland soils with a range of historical moisture regimes' for processing publication data downloads.
Category
Data Science & Biostatistics
Category
Dr. Paul Piehowski is the Proteomics team leader for PNNL’s Environmental and Molecular Sciences Division and the Environmental Molecular Sciences Laboratory (EMSL) user program. Piehowski is an analytical chemist whose research is focused on the application of mass spectrometry to biological...
Category
Viral communities detected from three large grassland soil metagenomes with historically different precipitation moisture regimes.
Category
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from KPBS field site in Manhattan, KS. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.