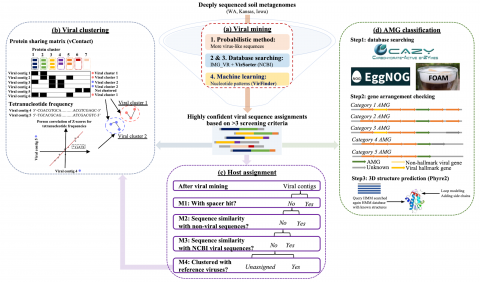
Viral genome assembly annotations from terabase metagenomes (TmG.1.0) from uncultivated soil collections from the IAREC field sites in Washington (IAREC), Kansas (KBPS), and Iowa (COBS), USA. Uncultivated virus Genomes (identified from soil metagenome datasets) Data DOI Package.
Filter results
Category
- (-) Earth System Science (89)
- Scientific Discovery (178)
- Biology (106)
- Human Health (69)
- Integrative Omics (52)
- Microbiome Science (16)
- National Security (14)
- Computing & Analytics (10)
- Energy Resiliency (7)
- Computational Research (6)
- Visual Analytics (6)
- Atmospheric Science (3)
- Coastal Science (3)
- Computational Mathematics & Statistics (3)
- Data Analytics & Machine Learning (3)
- Data Analytics & Machine Learning (3)
- Renewable Energy (3)
- Chemical & Biological Signatures Science (2)
- Cybersecurity (2)
- Distribution (2)
- Ecosystem Science (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Weapons of Mass Effect (2)
- Bioenergy Technologies (1)
- Chemistry (1)
- Computational Mathematics & Statistics (1)
- Energy Efficiency (1)
- Energy Storage (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Plant Science (1)
- Solar Energy (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Soil Microbiology (13)
- sequencing (9)
- Metagenomics (7)
- Microbiome (4)
- Omics (4)
- Fungi (3)
- Genomics (3)
- IAREC (3)
- metagenomics (3)
- Sequencing (3)
- soil microbiology (3)
- Climate Change (2)
- Mass Spectrometry (2)
- omics (2)
- Amplicon Sequencing (1)
- Chitin (1)
- High Throughput Sequencing (1)
- Metatranscriptomic (1)
- microbiome stability (1)
- Microscopy (1)
- mineral weathering (1)
- MPLEX (1)
- MultiSector Dynamics (1)
- Output Databases (1)
- Software Data Analysis (1)
- Soil (1)
- species volatility (1)
- Spectroscopy (1)
- Viruses (1)
- Whole Genome Sequencing (1)
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from KPBS field site in Manhattan, KS. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from COBS field site in Boone County, IA. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from IAREC field site in Prosser, WA. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
The data accompanies the manuscript in review that evaluates salinity-associated shifts in organic C thermodynamics, biochemical transformations, and heteroatom content in a first-order coastal watershed in the Olympic Peninsula of Washington state, USA. The files contain raw data including soil...
Category
The data accompanies the manuscript that evaluates salinity-associated shifts in organic C thermodynamics, biochemical transformations, and heteroatom content in a first-order coastal watershed in the Olympic Peninsula of Washington state, USA. The files contain raw data including soil chemical...
Category
The data accompanies the manuscript in review that evaluates salinity-associated shifts in organic C thermodynamics, biochemical transformations, and heteroatom content in a first-order coastal watershed in the Olympic Peninsula of Washington state, USA. The files contain raw data including soil...