Influenza A Experiment IM104 Processed Omics Data Unavailable This Influenza experiment evaluated mouse lung expression after etoposide treatment and infection with a pandemic H1N1 influenza strain. Related Experimental Data BioProject: PRJNA382278 GEO: GSE97555 (mRNA transcriptome response)...
Filter results
Content type
Tags
- (-) Influenza A (9)
- (-) Genomics (4)
- Virology (63)
- Immune Response (48)
- Time Sampled Measurement Datasets (45)
- Gene expression profile data (42)
- Differential Expression Analysis (41)
- Homo sapiens (30)
- Mass spectrometry data (25)
- Multi-Omics (23)
- MERS-CoV (16)
- Mus musculus (15)
- Viruses (14)
- Health (13)
- Soil Microbiology (13)
- Virus (13)
- West Nile virus (11)
- Synthetic (10)
- Ebola (9)
- sequencing (9)
- Resource Metadata (8)
- Metagenomics (7)
- Microarray (5)
- Human Interferon (4)
- Microbiome (4)
- Omics (4)
- IAREC (3)
- metagenomics (3)
- Sequencing (3)
- soil microbiology (3)
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM101 The purpose of this experiment was to evaluate the host mouse response to Influenza A virus (subtype H5N1) wild-type strain Influenza A/Vietnam/1203/2004, Influenza A/Vietnam/1203/2004 mutant strains PB2-627E...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM102 The purpose of this experiment was to evaluate the mouse host response to Influenza A virus (subtype H7N9) wild-type strain Influenza A/Anhui/1/2013 (AH1-WT) virus and mutants NS1-103F/106M (AH1-F/M) and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM103 The purpose of this experiment was to evaluate the host response to Influenza A virus (subtype H5N1) wild-type strain Influenza A/Vietnam/1203/2004 (VN1203) virus, mutant VN1203-NS1trunc124, and mock...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL103 The purpose of this experiment was to evaluate the human host response to Influenza A virus (subtype H5N1) and mutant viruses. Sample data was obtained for human lung adenocarcinoma cell line (Calu-3)...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL104 The purpose of this experiment was to evaluate the host response to pandemic Influenza A virus (subtype H1N1) , natural isolate Influenza A/California/04/2009 virus. Sample data was obtained from human lung...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL102 The purpose of this experiment was to evaluate the human host cellular response to Influenza A virus (subtype H7N9) , wild-type strain Influenza A/Anhui/1/2013 (AH1-WT), and mutant viruses NS1-L103F/I106M...
Category
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL105 Metadata The purpose of this experiment was to evaluate the host epigenetic response to Influenza A virus (subtype H5N1) infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL106 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to Influenza A virus (subtype H5N1) infection. Samples were obtained from human lung adenocarcinoma cell line (Calu...
Category
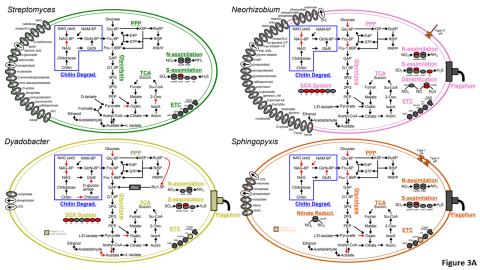
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes. [Data Set] PNNL DataHub. https://doi.org/10.25584/PNNLDH/1986536 Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes...
Category
Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...