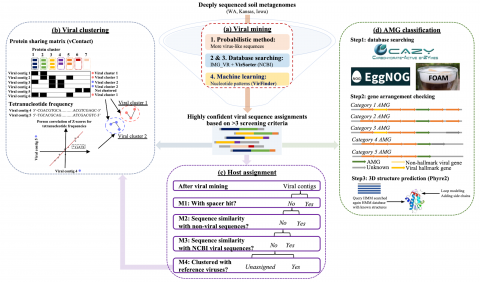
The following R source code was used for plotting figures of the viral communities detected from three grasslands soil metagenomes with a historical precipitation gradient ( WA-TmG.2.0 , KS-TmG.2.0 , IA-TmG.2.0 ) from project publication 'DNA viral diversity, abundance and functional potential vary...
Filter results
Showing 1 - 6 of 6
Fusarium sp. DS682 Proteogenomics Statistical Data Analysis of SFA dataset download: 10.25584/KSOmicsFspDS682/1766303 . GitHub Repository Source: https://github.com/lmbramer/Fusarium-sp.-DS-682-Proteogenomics MaxQuant Export Files (txt) Trelliscope Boxplots (jsonp) Fusarium Report (.Rmd, html)...
Code pertaining to the Soil Microbiome SFA Project publication data visualizations 'DNA viral diversity, abundance and functional potential vary across grassland soils with a range of historical moisture regimes' for processing publication data downloads.
Category
The srpAnalytics modeling pipeline for the Superfund Research Program Analytics Portal.
Category
Guide
Guide
1. Select the login button, where you will be prompted to login to ORCID 2. If you already have an ORCID, you may sign into it here and proceed. If not, please create an account. 3. DataHub needs the ORCID email visibility settings to be open, so next we'll need to change those settings in ORCID. If...