Description
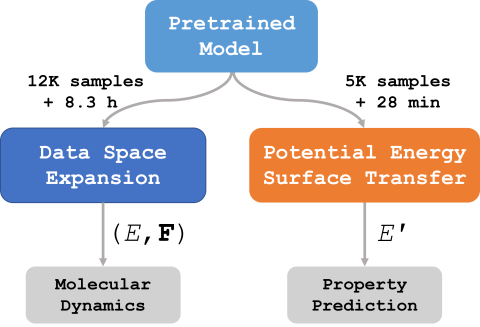
The demonstrated success of transfer learning has popularized approaches that involve pretraining models from massive data sources and subsequent finetuning towards a specific task. While such approaches have become the norm in fields such as natural language processing, implementation and evaluation of transfer learning approaches for chemistry are in the early stages. In this work, we demonstrate finetuning for downstream tasks on a graph neural network (GNN) trained over a molecular database containing 2.7 million water clusters. The use of Graphcore IPUs as an AI accelerator for training molecular GNNs reduces training time from a reported 2.7 days on 0.5M clusters to 1.2 hours on 2.7M clusters. Finetuning the pretrained model for downstream tasks of molecular dynamics and transfer to a different potential energy surface took only 8.3 hours and 28 minutes, respectively, on a single GPU.
The accompanying codebase is located at https://github.com/pnnl/downstream_mol_gnn.