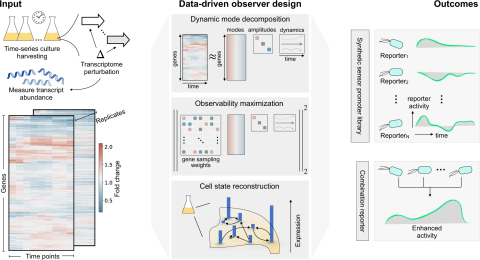
A major challenge in biotechnology and biomanufacturing is the identification of a set of biomarkers for perturbations and metabolites of interest. Here, we develop a data-driven, transcriptome-wide approach to rank perturbation-inducible genes from time-series RNA sequencing data for the discovery of analyte-responsive promoters. This provides a set of biomarkers that act as a proxy for the transcriptional state referred to as cell state. We construct low-dimensional models of gene expression dynamics and rank genes by their ability to capture the perturbation-specific cell state using a novel observability analysis. Using this ranking, we extract 15 analyte-responsive promoters for the organophosphate malathion in the underutilized host organism Pseudomonas fluorescens SBW25. We develop synthetic genetic reporters from each analyte-responsive promoter and characterize their response to malathion. Furthermore, we enhance malathion reporting through the aggregation of the response of individual reporters with a synthetic consortium approach, and we exemplify the library’s ability to be useful outside the lab by detecting malathion in the environment. The engineered host cell, a living malathion sensor, can be optimized for use in environmental diagnostics while the developed machine learning tool can be applied to discover perturbation-inducible gene expression systems in the compendium of host organisms.
Linked Primary Data
Sequencing
- Reference Taxonomy: Pseudomonas fluorescens (strain SBW25)
- NCBI BioProject: PRJNA826870
- NCBI GEO: GSE200822
Source Code
Reference Citation
Hasnain, A., Balakrishnan, S., Joshy, D.M. et al. Learning perturbation-inducible cell states from observability analysis of transcriptome dynamics. Nat Commun 14, 3148 (2023). https://doi.org/10.1038/s41467-023-37897-9
Last updated on 2023-07-26T15:12:26+00:00 by LN Anderson
Projects (1)
Last updated on 2023-02-23T19:37:46+00:00 by LN Anderson PerCon SFA Project Publication Experimental Data Catalog The Persistence Control of Engineered Functions in Complex Soil Microbiomes Project (PerCon SFA) at Pacific Northwest National Laboratory ( PNNL ) is a Genomic Sciences Program...
Datasets
5