Category
Description
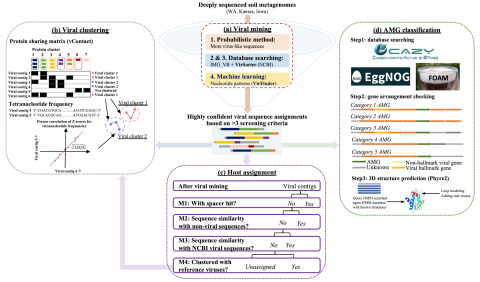
Soil samples were collected in triplicate in the Fall of 2017 across the three grassland locations having differences in historical annual precipitation (WA-TmG.1.0, KS-TmG.1.0, and IA-TmG.1.0). For each location, one deeply sequenced composite metagenome (> 1 terabase) were screened for viruses,In addition, each of the three field soil samples were individually sequenced to provide three replicate metagenomes (WA-TmG.2.0, KS-TmG.2.0, and IA-TmG.2.0) for statistical comparison in addition to screening for viral soil contigs for assessing the impact of historical annual precipitation. A total of 2,631 viral contigs were identified from the three grassland soil metagenomes, resulting in 14 newly completed high-quality viral genomes. The total number of viral contigs was highest in WA (1,577 contigs), followed by KS (756 contigs), and lowest in IA (298 contigs). This data package provides new knowledge of viral types, abundances, diversity, and auxiliary metabolic genes (AMGs) in grassland soils for predicting viral response to changes in their environment.
Viral genome sequencing identified from a metagenome and/or metatranscriptome dataset. Viral genomes are the result of computational analysis where metagenome datasets contained mixed genetic material information from sample sequences recovered directly from the natural environment. The version described in this paper is the first version. Dataset download contains functional annotations, computational analysis files, and a MIUVIG.soil.5.0 metadata file.
Data Package Resource Files:
- Iso-VIG14.1.0 MIUVIG.soil.5.0
- https://github.com/hafen/trelliscopejs
- https://github.com/Ruonan0101/GrasslandVirome_PNNL
Items below available from Download Button:
- 14_complete_V_genome.fasta (WA & KS only) [1.2 MB;1 item]
- Supporting Information Material [13.4 MB; 8 items]
- SI1 Differences in soil chemistry of the grasslands across precipitation gradient
- SI2 Prokaryotic community compositions of the grasslands across precipitation gradient
- SI3 Host assignments and clustering information of each identified viral contigs
- SI4 Visualization of the complete viral genomes assembled from soil metagenomes
- SI5 Statistical information of the ‘large’ (TmG.1.0) and ‘replicate’ (TmG.2.0) metagenomes
- SI6 Viral Pfam list for viral gene annotation
- SI7 Network of viral clustering using tetranucleotide frequency method
- SI8 An interactive network of the viral clustering (html)
Related Experimental Data
TmG.1.0 Metagenomes: 10.25584/WATmG1/1635002 | 10.25584/KSTmG1/1635004 | 10.25584/IATmG1/1635005
TmG.2.0 Metagenomes: 10.25584/WATmG2/1770324 | 10.25584/KSTmG2/1770332 | 10.25584/IATmG2/1770333