Description
Genomic DNA was isolated from soil and rhizosphere samples using the Zymo Quick-DNA fecal/soil microbe miniprep kit (catalog no. D6010) according to the manufacturer’s instructions (Zymo Research; Irvine, CA) with the modification of eluting in 100 uL elution buffer. Sample concentrations were quantified using the Qubit dsDNA HS assay kit (Thermo Fisher). For rhizosphere samples only, DNA was subsequently purified using Zymo’s ZR-96 DNA Clean & Concentrator kit (catalog no. D4024) to account for low DNA concentrations of these samples. . In each replicate block, there were five drip irrigation treatments (T1 = 100% normal irrigation, T2 = 56.25%, T3 = 37.5%, T4 = 18.75% and T5=no irrigation. On July 20, the strength of the drought treatments was increased: T1 remained at 100%, whereas T2 changed from 75% to 56.25%, T2 changed from 50% to 37.5%, and T4 changed from 25% to 18.75%.
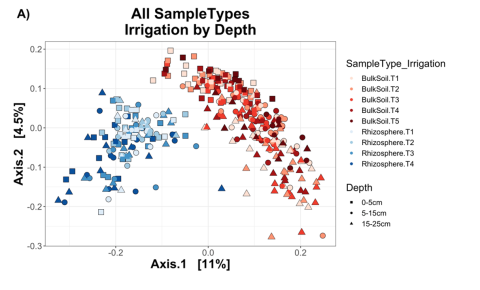
Sequencing was performed as described previously (Naylor, Fansler, et al. 2020). Sequences were amplified on the MiSeq platform (Illumina, San Diego, CA) using 16S primers (515F and 806R) specific to the V4 region. Raw sequence data was processed with the pipeline Hundo for amplicon quality control and annotation. Downstream statistical analyses on 16S datasets were performed using the program R and the packages ‘phyloseq’ and ‘vegan’.