Data Sample
Description
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson
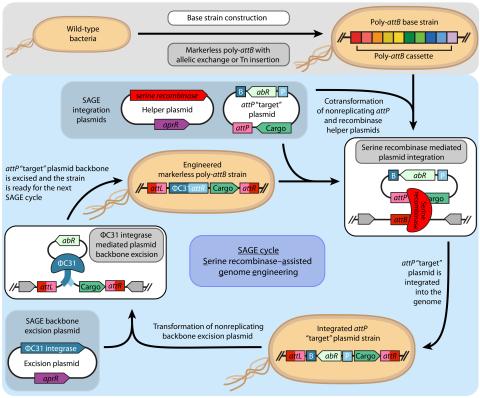
SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis
Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project.
Raw Measurement Data
- NCBI BioProject: PRJNA841687 (15 BioSamples; 120 raw sequence reads)
Related Organisms
- Rhodopseudomonas palustris CGA009 (NCBITaxon:258594)
- Pseudomonas putida GPO1 (NCBITAXON:303)
- Rhodococcus jostii RHA1 (NCBITAXON:132919)
- Pseudomonas frederiksbergensis TBS10 (NCBITAXON:104087)
- Pseudomonas fluorescens SBW25 (NCBITAXON:294)
Reference Citations
- Elmore JR, Dexter GN, Baldino H, Huenemann JD, Francis R, Peabody GL 5th, Martinez-Baird J, Riley LA, Simmons T, Coleman-Derr D, Guss AM, Egbert RG. High-throughput genetic engineering of nonmodel and undomesticated bacteria via iterative site-specific genome integration. Sci Adv. 2023 Mar 10;9(10):eade1285. doi: https://doi.org/10.1126/sciadv.ade1285
- Robert G. Egbert, & Joshua R. Elmore. (2022). Serine Recombinase-Assisted Genome Engineering - Relative Transcriptional Profiling (SAGE-RTP) Toolkit (1.0). Zenodo. https://doi.org/10.5281/zenodo.7388406
Funding Acknowledgments
The provided material is based upon work supported by the U.S. Department of Energy, Office of Biological and Environmental Research, Genomic Science Program and is a contribution of the Pacific Northwest National Laboratory Persistence Control of Engineered Functions in Complex Soil Microbiomes project. Pacific Northwest National Laboratory is a multiprogram national laboratory managed by the Battelle Memorial Institute Battelle Memorial Institute, operating under the U.S. Department of Energy Contract DE-AC05-76RL01830.
Data Reuse Citation Policy
When referencing and reusing linked PerCon SFA project digital data assets listed at the PNNL DataHub institutional repository, we ask that users please cite the dataset digital object identifier provided at the download page and any corresponding journal article publications. Reference citations, where applicable, should provide the necessary metadata information required to support, corroborate, verify, and otherwise determine the legitimacy of the research findings provided (data and code) from scholarly publications and corresponding project data releases.
Data Licensing
CC BY-SA 4.0 (processed datasets), CC0 1.0 PNNL DataHub policy default for metadata)