Category
Description
Created 2025-06-03T23:16:59+00:00 by LN Anderson.Currently pending DOI assignment for public release.
Integrative SP3 Workflow for Multi-PTM Proteomics Profiling (PPI-TZ-DP0)
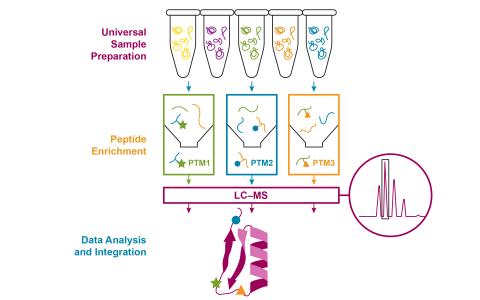
The purpose of this experiment was to evaluate a new standardized method approach for proteomics sample processing designed to facilitate multiplexed quantification of protein abundance, cysteine thiol oxidation, phosphorylation, and acetylation starting from the same sample material. Experimental samples derived from alveolar type II lung epithelial cells (C10 cells), BALB/C mouse gastrocnemius muscle tissue, and cytokine cocktail time course treated β-TC-6 cells (4, 8, and 24 hrs.) were prepared using established automated multi-PTM SP3 bead enrichment and cleanup workflow, followed by fractionation for TMT quantitation analysis or label-free quantitation analysis.
Accessible Digital Data Downloads
Proteomic data was acquired using a Q-Exactive Plus mass spectrometer or a Vanquish Neo UHPLC coupled with an Orbitrap Exploris 480 mass spectrometer and processed using the PlexedPiper software workflow and MaxQuant software. Processed datasets are openly accessible from the download button and contain processed global proteomic, redox proteomic, and phosphoproteomic data results files and experimental design metadata. Processed data package download includes sample naming keys, raw abundance files, sequence annotations, and supporting metadata materials. See corresponding primary data accessions below and linked source code in supporting data transparency and reuse.
Linked Primary Data Accessions
Primary liquid chromatography-mass spectrometry (LC-MS) raw measurement proteome data are openly accessible for download at the Mass Spectrometry Interactive Virtual Environment (MassIVE) community repository under the accession MSV000095264.
Data Download Reference Citations
- Gluth A, Li X, Gritsenko MA, Gaffrey MJ, Kim DN, Lalli PM, Chu RK, Day NJ, Sagendorf TJ, Monroe ME, Feng S, Liu T, Yang B, Qian WJ, Zhang T. Integrative Multi-PTM Proteomics Reveals Dynamic Global, Redox, Phosphorylation, and Acetylation Regulation in Cytokine-Treated Pancreatic Beta Cells. Mol Cell Proteomics. 2024 Dec;23(12):100881. doi: 10.1016/j.mcpro.2024.100881. Epub 2024 Nov 15. PMID: 39550035; PMCID: PMC11700301.
Funding Acknowledgments
The research data described here was funded in whole or in part by the Predictive Phenomics Initiative (PPI) at Pacific Northwest National Laboratory (PNNL). This work was conducted under the Laboratory Directed Research and Development Program at PNNL. A portion of this research was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy (DOE) Office of Science located at PNNL. PNNL is a multiprogram national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RL01830.
Citation Policy
In efforts to enable discovery, reproducibility, and reuse of PPI-funded project dataset citations in accordance with best practices (as outlined by the FORCE11 Data Citation Principles), we ask that all reuse of project data and metadata download materials acknowledge all primary and secondary dataset citations and corresponding journal articles where applicable.
Data Licensing