Category
Description
Predictive Phenomics Initiative (PPI) Project Data Catalog Collection
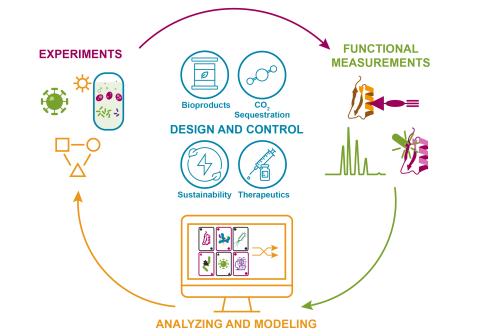
The Predictive Phenomics Initiative (PPI) is an internal LDRD investment at Pacific Northwest National Laboratory focused on unraveling the mysteries of molecular function in complex biological systems. Explore PPI research project data package downloads, containing processed phenomics data results (and curated experimental metadata) linked to raw measurement data, software/source code analysis deliverables, and corresponding peer-reviewed publications where applicable.
PPI Research Project Thrust Areas
- TA1 - Enhancing Multi-Scale Phenomics Measurements
- TA2 - Science Drivers for Collaboration
- TA3 - Computational Methods for Predicting & Controlling the Phenome
For a complete catalog of available project software/source code, supporting phenomics data analysis and reuse, visit the PPI GitHub Repository and Zenodo Community Collection.
Funding Acknowledgments
The research data described here was funded by the Predictive Phenomics Science & Technology Initiative (PPI), conducted under the Laboratory Directed Research and Development Program, at Pacific Northwest National Laboratory (PNNL). PNNL is a multiprogram national laboratory, operated by Battelle, U.S. Department of Energy, Office of Science under Award Number DE-AC05-76RL01830.
Citation Policy
In efforts to enable discovery, reproducibility, and reuse of PPI-funded project dataset citations in accordance with best practices (e.g., FORCE11 Data and Software Citation Principles), we ask that all reuse of project data packages and linked materials acknowledge all primary and secondary dataset citations, source code citations, and corresponding peer-reviewed journal articles where applicable.
Data Licensing
Creative Commons Attribution 4.0 International (CC BY 4.0)
Last updated on 2025-07-14T18:56:42+00:00 by LN Anderson
Projects (10)
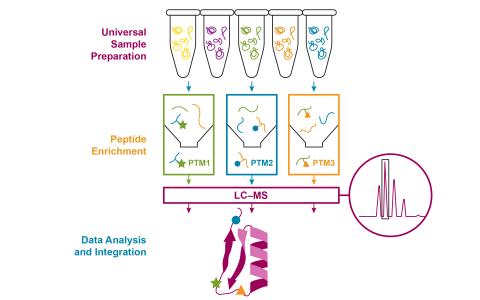
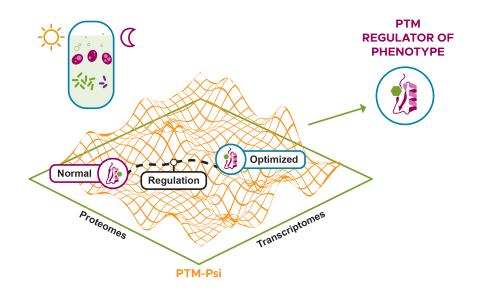
The research objective of this project is to develop an integrative and automated multi-PTM profiling capability with deep proteome coverage.
Category
Datasets
2
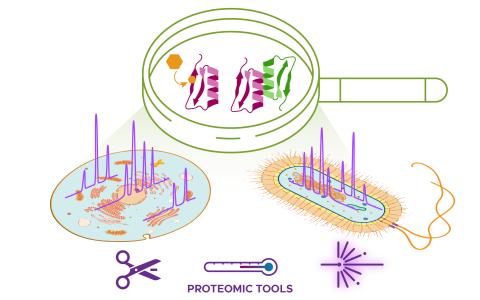
The science objective of this project is to apply structural proteomics technologies to map the molecular interactome.
Category
Datasets
5
The science objectives of this project are to: Functionally enrich microbial communities and generate multi-omics to correlate biochemical mechanisms to activity. Integrate PhenoProfiling with Thrust Areas 2 and 3 to develop models for phenotype prediction and interspecies interactions. Evaluate...
Category
Datasets
1
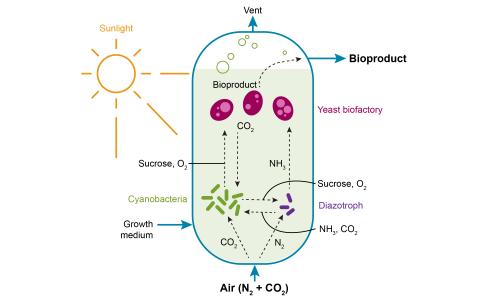
The research goal of this project is to establish model synthetic microbial communities to understand the rules regulating their biological function in order to utilize them as next generation bioproduction platforms capable of reducing carbon and nitrogen footprints in biomanufacturing processes.
Category
Datasets
4
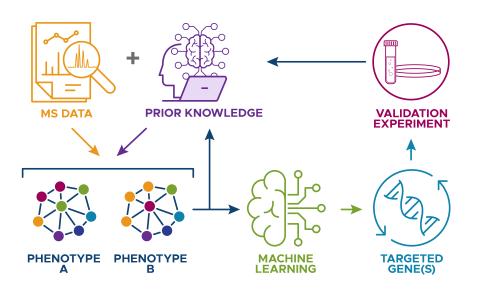
The research goal of this project is to develop a biologically informed machine learning (ML) model that integrates datasets from different studies, and leverages current biological knowledge in an automated manner, to improve predictions in biological data analysis.
Category
Datasets
0
The research goal of this project is to develop new theory and tools that leverage evolutionary perspectives and knowledge of the energetics of reactions to predict the most likely regulation in a given environment. These methods will accelerate exploration, modeling and understanding of cell...
Category
Datasets
3
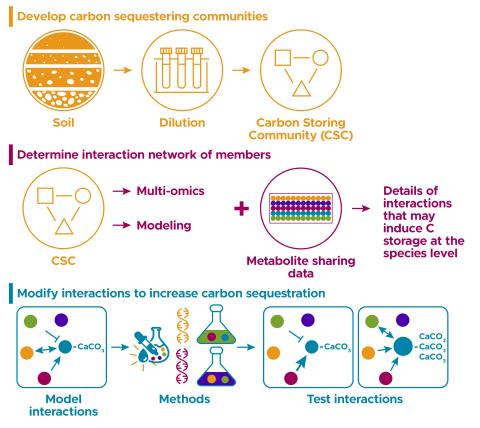
The research goal of this project is to build and understand model communities that show carbon storage phenotypes
Category
Datasets
0
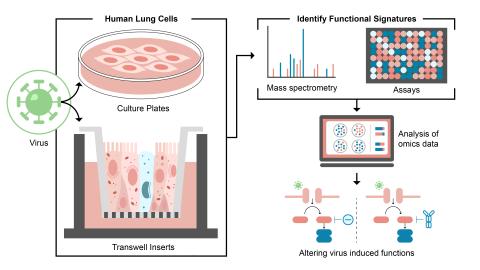
This project will improve current understanding of how viruses manipulate host environments. Use cutting edge, iterative proteomics and metabolomics tools in ex vivo primary human lung cultures that recapitulate the epithelium of the conducting airway and novel high throughput sample capture...
Datasets
5
The research goal of this project is to develop a computational approach known as Variation-leveraged Phenomic Association Study (VaLPAS) to address the challenge of using functional dark matter (proteins, metabolites, lipids) for bioeconomy applications. The project has four objectives: 1...
Category
Datasets
0
The overarching goal of this research is to predict and engineer robust microbial phenotypes under stressed conditions to accelerate and de-risk bioprocess development. Laboratory strains routinely fail to maintain productivity at industrial scales, in part due to bioprocess stresses and a limited...
Category
Datasets
0