Category
Description
Dataset Description
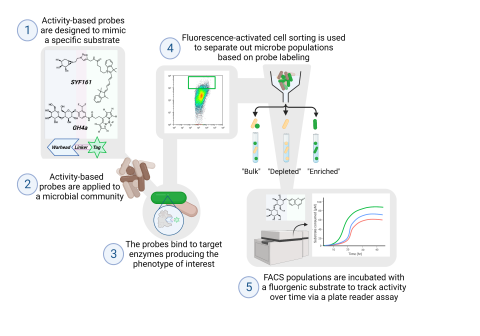
Soil samples were incubated with either 0.2% cellobiose for three days or 0.2% xylan for nine days. Microbes were extracted from the soil matrix, incubated overnight in MOPS buffer with either cellobiose or xylose. The following day subsamples from each carbon source incubations were labeled with activity-based probes. For the cellobiose incubated sample, the GH4a probe was used to target glucosidase and cellobiosidase activity, and for the xylan incubated samples, the SYF161 probe was used to target xylosidase and xylobiosidase activity. The GH4a probe was conjugated to a BODIPY (excitation = 502 nm, emission = 511 nm) tag requiring Syto59 (excitation = 622 nm, emission = 645 nm) to be used as the general nucleic acid stain, while SYF161 was conjugated to a Cy5 (excitation = 651 nm, emission = 670 nm) tag and coupled with Syto9 (excitation = 485 nm, emission = 498 nm). The total microbial population, probe unlabeled, and probe labeled were separated based on fluorescence intensity via fluorescence-activated cell sorting (FACS). Samples were collected for direct 16S rRNA sequencing. Additional samples were collected for further incubations in MOPS buffer with either cellobiose or xylose. Aliquots were taken during the post-FACS incubations to track the growth of microbial communities via 16S gene sequencing. Additional negative controls of reagents were collected throughout the process. All samples were sequenced targeting the v4 region of the 16S gene and sequenced using Illumina MiSeq v2 chemistry on a 2x250 flow cell.
Accessible Digital Data Downloads
The repository contains the following files:
- ACS-NR-DP1_SampleMetadata.xlsx: Contains sample metadata information including descriptors, experimental conditions, cell lines (if applicable)
- ACS-NR-DP1_Amplicon.xlsx: Contains the 16S amplicon counts for all samples
Total Download Size: 4.6 MB, zipped
Linked Publications
Reichart NJ, Steiger AK, Van Fossen EM, McClure R, Overkleeft HS, Wright AT. Selection and enrichment of microbial species with an increased lignocellulolytic phenotype from a native soil microbiome by activity-based probing. ISME Commun. 2023 Sep 30;3(1):106. doi.org/10.1038/s43705-023-00305-w
Linked Primary Data
Primary raw measurement data are openly accessible for download at the Gene Expression Omnibus (GEO) community repository under the accession: <not yet submitted>
Funding Acknowledgments
The research data described here was funded in whole or in part by the Predictive Phenomics Initiative (PPI) at Pacific Northwest National Laboratory (PNNL). This work was conducted under the Laboratory Directed Research and Development Program at PNNL. A portion of this research was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy (DOE) Office of Science located at PNNL. PNNL is a multiprogram national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RL01830.
Citation Policy
In efforts to enable discovery, reproducibility, and reuse of PPI-funded project dataset citations in accordance with best practices (as outlined by the FORCE11 Data Citation Principles), we ask that all reuse of project data and metadata download materials acknowledge all primary and secondary dataset citations and corresponding journal articles where applicable.
Data Licensing
Creative Commons Attribution 4.0 International (CC BY 4.0)