Category
Description
Dataset Description
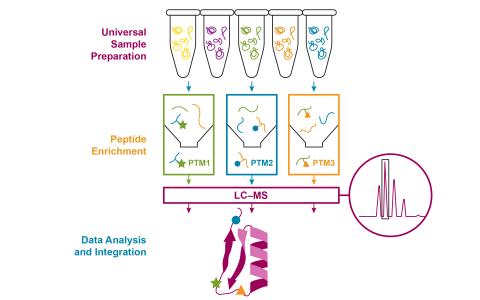
The purpose of this experiment was to evaluate the regulatory stress response of Oleaginous yeast species Rhodotorula toruloides NBRC 0880 (JGI strain IFFO0880 v4.0) under nitrogen-rich and nitrogen-limited conditions over time. Time course experimental samples (24, 48, and 72 hours after inoculation) were prepared using a semi-automated multi-PTM proteomic approach, using tandem mass tag 18-plex (TMT18), and lipidome remodeling for downstream multi-omics analysis.
Processed datasets are openly accessible from the download button and contain secondary processed proteomic (redox, phospho, and global TMT) and lipidomic (positive and negative ion mode) results files and experimental design metadata. Experimental proteomic sample data was acquired using an Orbitrap Exploris 480 mass spectrometer and processed and compiled using PlexedPiper software workflow and custom R. toruloides Nitrogen Limitation Multi-PTM Profiling R Package (located at Zenodo). Experimental lipidomic sample data were collected in parallel with proteomic samples and acquired using an Orbitrap Fusion Lumos mass spectrometer and further processed using MS-DIAL, lipidr R package, and MetaboAnalyst (v.6.0) software. Processed data package download includes sample naming keys, raw abundance files, sequence annotations, and supporting metadata materials. See corresponding primary data accessions below and linked source code in supporting data transparency and reuse.
Data Download Reference Citation
Gluth, Austin; Czajka, Jeffrey J; Li, Xiaolu; Bloodsworth, Kent J; Eder, Josie G; Kyle, Jennifer E; Chu, Rosalie K; Yang, Bin; Qian, Wei-Jun; Bohutskyi, Pavlo; Waters, Katrina M; & Zhang, Tong. (2025) Rhodotorula toruloides Nitrogen Limitation PTM Profiling Multi-Omics (TZ-DP1). https://doi.org/10.25584/2510803
Accessible Digital Data Downloads
The repository contains the following folders and files:
- TZ-DP1_SampleMetadata_Lipidomics.xlsx: Contains sample metadata information including descriptors, experimental conditions, cell lines (if applicable)
- TZ-DP1_ExpressionData_Lipidomics.xlsx: Contains raw abundances and statistics for lipidomics data of Rhodotorula toruloides
- TZ-DP1_SampleMetadata_Proteomics.xlsx: Contains sample metadata information including descriptors, experimental conditions, cell lines (if applicable)
- TZ-DP1_ExpressionData_Proteomics.xlsx: Contains raw abundance and statistics for proteomics data and PTMs of Rhodotorula toruloides
Total Download Size: 5.8 MB, zipped
Linked Primary Data Accessions
Primary liquid chromatography-mass spectrometry (LC-MS) raw measurement proteome data are openly accessible for download at the Mass Spectrometry Interactive Virtual Environment (MassIVE) community repository under the accession MSV000095798. Primary LC-MS raw measurement lipidome data (positive and negative mode) are openly accessible for download at MassIVE under the accession MSV000096720.
Funding Acknowledgments
The research data described here was funded in whole or in part by the Predictive Phenomics Initiative (PPI) at Pacific Northwest National Laboratory (PNNL). This work was conducted under the Laboratory Directed Research and Development Program at PNNL. A portion of this research was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy (DOE) Office of Science located at PNNL. PNNL is a multiprogram national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RL01830.
Citation Policy
In efforts to enable discovery, reproducibility, and reuse of PPI-funded project dataset citations in accordance with best practices (as outlined by the FORCE11 Data Citation Principles), we ask that all reuse of project data and metadata download materials acknowledge all primary and secondary dataset citations and corresponding journal articles where applicable.
Data Licensing