Category
Description
Please cite as: Robert E. Danczak, Sheryl Bell, Evan Warburton, Trinidad Alfaro, Kirsten Hofmockel, Amy E. Zimmerman, William C. Nelson, 2025. Prokaryotic and eukaryotic interactions along a moisture gradient. [Data Set] PNNL DataHub. https://doi.org/10.25584/2574929
This data is published under a CC0 license. The authors encourage data reuse and request attribution by referencing the above citations for the data package and associated manuscript.
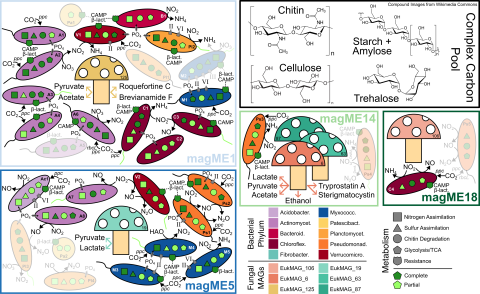
We generated genome-resolved multiomics data from a series of metagenomic and metatranscriptomic sequencing. Specifically, we acquired, functionally annotated, and taxonomically classified both bacterial and eukaryotic metagenome assembled genomes (MAGs). For bacterial MAGs, we assembled eukaryotic float metagenomic sequencing data from JGI using MEGAHIT, binned and refined MAGs using MetaWRAP and dRep, functionally annotated MAGs using eggNOG mapper, and assigned taxonomy using GTDB-tk. For eukaryotic MAGs, we first identified potentially eukaryotic contigs from a coassembly of eukaryotic float metagenomic sequencing data from JGI using EukRep and Whokaryote, binned MAGs using MetaBAT2, functionally annotated MAGs using eggNOG mapper, and assigned taxonomy using Eukulele. Bulk metatranscriptomic reads were mapped to bacterial MAGs and polyA-metatranscriptomic read were mapped to eukaryotic MAGs using bbmap.