Category
Description
Please cite as: Josué A. Rodríguez-Ramos, Amy E. Zimmerman, Ruonan Wu, Sheryl Bell, Trinidad Alfaro, Kirsten Hofmockel, William C. Nelson. 2025. Inter-Kingdom Viral Interactions. [Data Set] PNNL DataHub. https://doi.org/10.25584/2583337
This data is published under a CC0 license. The authors encourage data reuse and request attribution by referencing the above citations for the data package and associated manuscript.
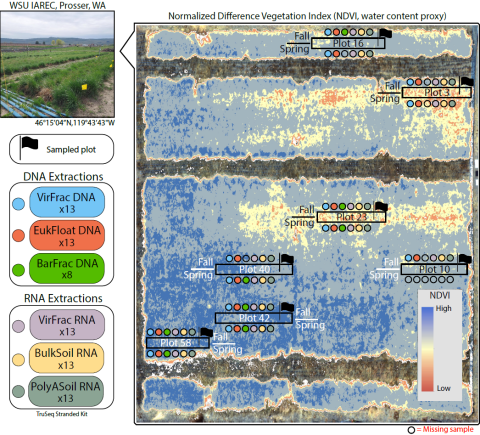
Deciphering viral ecology in soils is challenging due to their high physiochemical and community complexity. To enhance detection of sub-communities of DNA and RNA viruses, we applied fractionation approaches to soils collected across a moisture gradient from a grassland field experiment. Analyses included metagenomics and metatranscriptomics of size-fractionated extracellular viruses (i.e., DNA and RNA viromes), metagenomics of bacteria/archaea- or eukaryote-enriched samples, and whole soil metatranscriptomes with rRNA-depletion or polyadenylation enrichment. While RNA virome and whole soil RNA methods captured similar viral diversity, RNA viromes identified longer, higher-quality genomes. Further, we showed that significantly more DNA viruses were active in higher moisture than lower moisture samples, whereas responses by overall diversity vary by genome type (DNA versus RNA genomes). Finally, we demonstrate the power of fractionation approaches for identifying distinct viral communities that infect unique hosts, which has significant implications for ecological investigations, particularly related to interkingdom interactions.