Filter results
Category
- (-) Computational Research (23)
- (-) Coastal Science (4)
- (-) Ecosystem Science (4)
- Scientific Discovery (307)
- Biology (198)
- Earth System Science (136)
- Human Health (102)
- Integrative Omics (73)
- Microbiome Science (42)
- National Security (21)
- Computing & Analytics (14)
- Chemistry (10)
- Energy Resiliency (9)
- Data Analytics & Machine Learning (8)
- Materials Science (7)
- Visual Analytics (6)
- Chemical & Biological Signatures Science (5)
- Computational Mathematics & Statistics (5)
- Weapons of Mass Effect (5)
- Atmospheric Science (4)
- Renewable Energy (4)
- Data Analytics & Machine Learning (3)
- Plant Science (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Energy Storage (2)
- Grid Cybersecurity (2)
- Solar Energy (2)
- Bioenergy Technologies (1)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Type 1 Diabetes (6)
- Autoimmunity (5)
- Machine Learning (5)
- Biomarkers (4)
- Molecular Profiling (4)
- Mass spectrometry-based Omics (3)
- Predictive Modeling (3)
- Software Data Analysis (3)
- Alternative Splicing (2)
- DNA Sequence Analysis (2)
- Functional Annotation Analysis (2)
- Kmers (2)
- Mass Spectrometry (2)
- Omics (2)
- Python (2)
- RNA Sequence Analysis (2)
- Snakemake (2)
- Chitin (1)
- Data Analysis (1)
- Fish Detection (1)
- High-Performance Computing (1)
- Hydropower (1)
- Marine and Hydrokinetic (1)
- Mass Spectrometer (1)
- Metatranscriptomic (1)
- Output Databases (1)
- Soil (1)
- Statistics (1)
- ToF-SIMS (1)
- underwater video (1)
Category
Biography Kelly is a senior data scientist in the Computational Biology group within the Biological Sciences Division at Pacific Northwest National Laboratory (PNNL). After earning a MS in Biostatistics from the University of Washington in 2012, she worked at a cancer research company for two years...
David Degnan is a biological data scientist who develops bioinformatic and statistical pipelines for multi-omics data, specifically the fields of proteomics, metabolomics, and multi-omics (phenotypic) data integration. He has experience with top-down & bottom-up proteomics analysis, genomics &...
The IONTOF TOF.SIMS 5 data source is a time-of-flight secondary ion mass spectrometer and powerful surface analysis tool used to investigate scientific questions in biological, environmental, and energy research. Among the most sensitive of surface analysis tools, it uses a high-vacuum technique...
Category
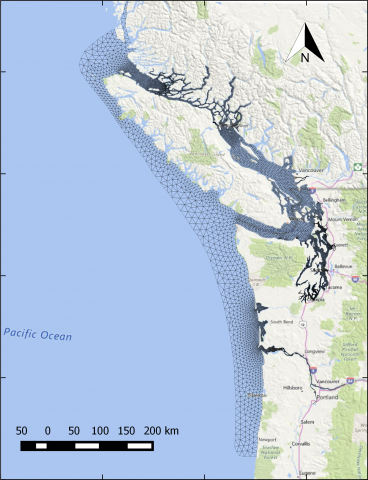
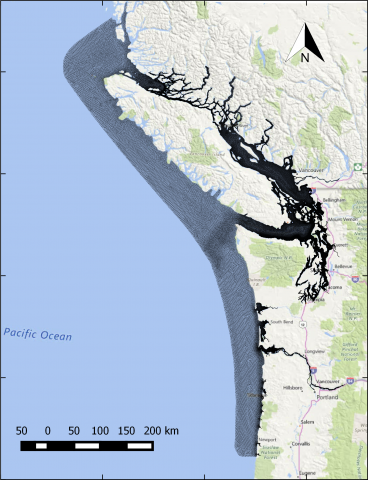
The year 2014 solution files are based on model calibration effort based on inputs and data from Year 2014 as described in Khangaonkar et al. 2018). It includes water surface elevation, currents, temperature and salinity at an hourly interval. These netcdf files with 24 hourly records include...
Category
The year 2014 solution files are based on model calibration effort based on inputs and data from Year 2014 as described in Khangaonkar et al. 2018). It includes water surface elevation, currents, temperature and salinity at an hourly interval. These netcdf files with 24 hourly records include...
Category
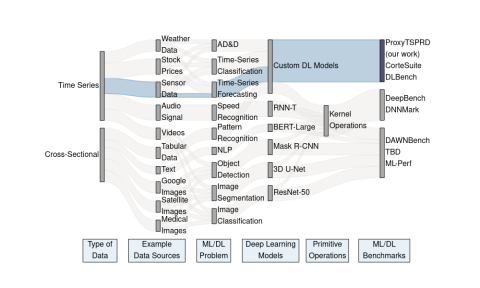
ProxyTSPRD profiles are collected using NVIDIA Nsight Systems version 2020.3.2.6-87e152c and capture computational patterns from training deep learning-based time-series proxy-applications on four different levels: models (Long short-term Memory and Convolutional Neural Network), DL frameworks...
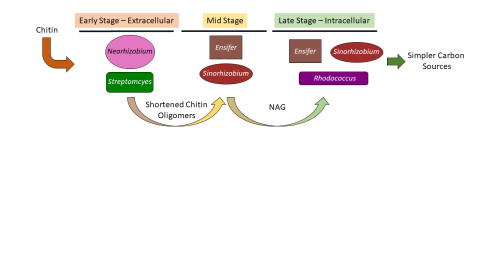
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Metatranscriptomic data from MSC-2. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33232 Metatranscriptomic data from MSC-2 12 fastq files (6 forward read, 6 reverse...
Category
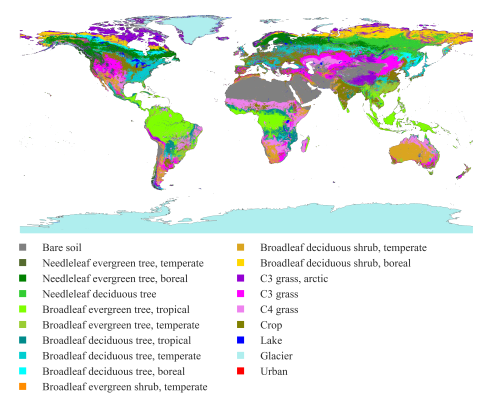
This dataset presents land surface parameters designed explicitly for global kilometer-scale Earth system modeling and has significant implications for enhancing our understanding of water, carbon, and energy cycles in the context of global change. Specifically, it includes four categories of...
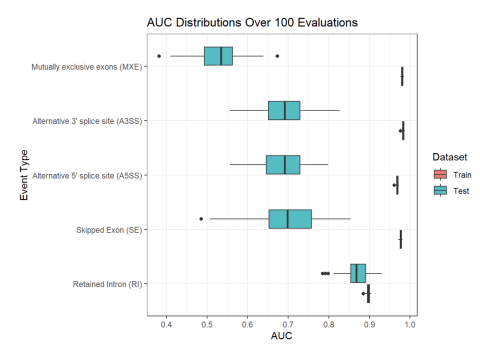
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...
A total of 172 children from the DAISY study with multiple plasma samples collected over time, with up to 23 years of follow-up, were characterized via proteomics analysis. Of the children there were 40 controls and 132 cases. All 132 cases had measurements across time relative to IA. Sampling was...
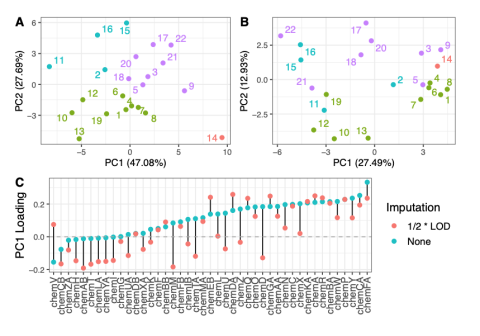
This data is supplementary to the manuscript Expanding the access of wearable silicone wristbands in community-engaged research through best practices in data analysis and integration by Lisa M. Bramer, Holly M. Dixon, David J. Degnan, Diana Rohlman, Julie B. Herbstman, Kim A. Anderson, and Katrina...
Comprised of 6,426 sample runs, The Environmental Determinants of Diabetes in the Young (TEDDY) proteomics validation study constitutes one of the largest targeted proteomics studies in the literature to date. Making quality control (QC) and donor sample data available to researchers aligns with...