Code pertaining to the Soil Microbiome SFA Project publication data visualizations 'DNA viral diversity, abundance and functional potential vary across grassland soils with a range of historical moisture regimes' for processing publication data downloads.
Filter results
Category
- (-) Biology (202)
- (-) Electric Grid Modernization (2)
- (-) Energy Efficiency (2)
- Scientific Discovery (311)
- Earth System Science (138)
- Human Health (102)
- Integrative Omics (72)
- Microbiome Science (44)
- Computational Research (25)
- National Security (22)
- Computing & Analytics (15)
- Chemistry (10)
- Energy Resiliency (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Materials Science (7)
- Visual Analytics (6)
- Chemical & Biological Signatures Science (5)
- Renewable Energy (5)
- Weapons of Mass Effect (5)
- Atmospheric Science (4)
- Coastal Science (4)
- Data Analytics & Machine Learning (4)
- Ecosystem Science (4)
- Plant Science (3)
- Cybersecurity (2)
- Distribution (2)
- Energy Storage (2)
- Grid Cybersecurity (2)
- Solar Energy (2)
- Bioenergy Technologies (1)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (50)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Multi-Omics (30)
- Viruses (27)
- Omics (25)
- Health (23)
- Virus (23)
- Soil Microbiology (22)
- MERS-CoV (18)
- Mus musculus (18)
- Mass Spectrometry (13)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- Ebola (9)
- High Throughput Sequencing (9)
- Microbiome (8)
- Proteomics (8)
- Microarray (7)
- Resource Metadata (7)
- Synthetic Biology (7)
- Imaging (6)
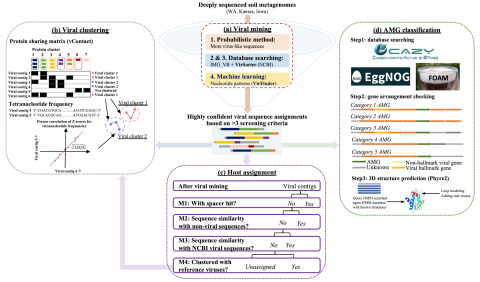
Viral communities detected from three large grassland soil metagenomes with historically different precipitation moisture regimes.
Category
Viral communities detected from three large grassland soil metagenomes with historically different precipitation moisture regimes.
Category
Exhaled breath condensate proteomics represent a low-cost, non-invasive alternative for examining upper respiratory health. EBC has previously been used for the discovery and validation of detected exhaled volatiles and non-volatile biomarkers of disease related to upper respiratory system distress...
Dr. Paul Piehowski is the Proteomics team leader for PNNL’s Environmental and Molecular Sciences Division and the Environmental Molecular Sciences Laboratory (EMSL) user program. Piehowski is an analytical chemist whose research is focused on the application of mass spectrometry to biological...
Category
The srpAnalytics modeling pipeline for the Superfund Research Program Analytics Portal.
Category
Citation: Gosline, S.J.C., Kim, D.N., Pande, P. et al. The Superfund Research Program Analytics Portal: linking environmental chemical exposure to biological phenotypes. Sci Data 10 , 151 (2023). https://doi.org/10.1038/s41597-023-02021-5 Funding Acknowledgments The research reported herein was...
Category
The srpAnalytics modeling pipeline for processing new data for the OSU-PNNL Superfund Research Program Analytics Portal located at https://github.com/sgosline/srpAnalytics Data License MIT License Copyright (c) 2020 Sara JC Gosline . Permission is hereby granted, free of charge, to any person...
Category
OSU-PNNL Superfund Research Program Center is part of the Superfund Research Program (SRP) at Oregon State University, directed by Dr. Robyn Tanguay, bringing together a multidisciplinary team of experts with extensive experience in polycyclic aromatic hydrocarbons (PAHs) research. Using state-of...
Category
Datasets
3