Dr. Bohutskyi’s research focus is in developing new bioprocesses addressing sustainable transformation of carbon dioxide, biomass, and wet or liquid waste, including carbon, nitrogen, phosphorus, and other critical elements into bioproducts, chemicals, and fuels. This research uses a suite of...
Filter results
Category
- Scientific Discovery (73)
- Biology (51)
- Human Health (16)
- Earth System Science (13)
- Integrative Omics (13)
- Computational Research (12)
- Microbiome Science (12)
- Chemistry (7)
- National Security (7)
- Data Analytics & Machine Learning (6)
- Computing & Analytics (5)
- Computational Mathematics & Statistics (3)
- Energy Resiliency (3)
- Chemical & Biological Signatures Science (2)
- Renewable Energy (2)
- Weapons of Mass Effect (2)
- Coastal Science (1)
- Data Analytics & Machine Learning (1)
- Ecosystem Science (1)
- Energy Efficiency (1)
- Energy Storage (1)
- Plant Science (1)
- Solar Energy (1)
Tags
- Predictive Phenomics (11)
- Mass Spectrometry (9)
- Multi-Omics (8)
- Virology (7)
- Omics-LHV Project (6)
- Differential Expression Analysis (5)
- Gene expression profile data (5)
- Immune Response (5)
- Time Sampled Measurement Datasets (5)
- Autoimmunity (4)
- Biomarkers (4)
- Homo sapiens (4)
- Mass spectrometry data (4)
- Molecular Profiling (4)
- Omics (4)
- Synthetic (4)
- Type 1 Diabetes (4)
- Machine Learning (3)
- Mass spectrometry-based Omics (3)
- Mus musculus (3)
- Proteomics (3)
- Synthetic Biology (3)
- Biological and Environmental Research (2)
- Ebola (2)
- Human Interferon (2)
- Influenza A (2)
- MERS-CoV (2)
- PerCon SFA (2)
- Predictive Modeling (2)
- West Nile virus (2)
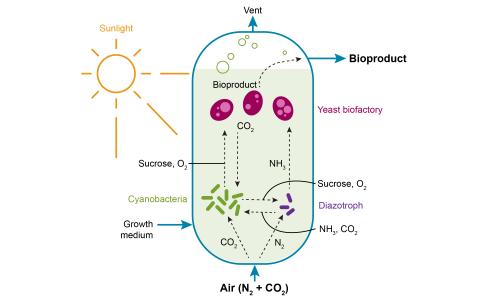
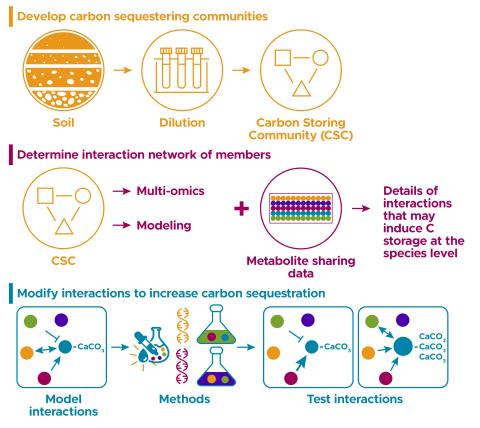
The research goal of this project is to establish model synthetic microbial communities to understand the rules regulating their biological function in order to utilize them as next generation bioproduction platforms capable of reducing carbon and nitrogen footprints in biomanufacturing processes.
Category
Datasets
0
Andrew McNaughton is a computational scientist currently working in the areas of modeling metabolic networks, multi-omics data analysis and integration, and computational chemistry. His areas of interest include high performance computing (HPC), artificial intelligence, and machine learning (ML) to...
Katherine joined the Sprunger Lab at the W. K. Kellogg Biological Station (part of Michigan State University) in January 2023. She came to KBS from Washington State University, where she earned her Ph.D. in soil science in 2022. Her research interests include agroecology with respect to how land...
Emeritus Professor Forage and Extension Agronomist at Washington State University https://css.wsu.edu/profile/?nid=fransen
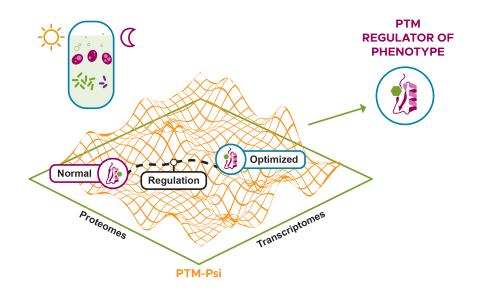
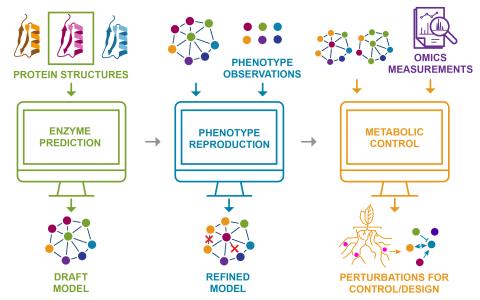
The research goal of this project is to develop computational methods to predict cell regulation phenotypes using small molecule and proteome data to understand outcomes in complex biological systems.
Category
Datasets
0
The research goal of this project is to develop new theory and tools that leverage evolutionary perspectives and knowledge of the energetics of reactions to predict the most likely regulation in a given environment. These methods will accelerate exploration, modeling and understanding of cell...
Category
Datasets
0
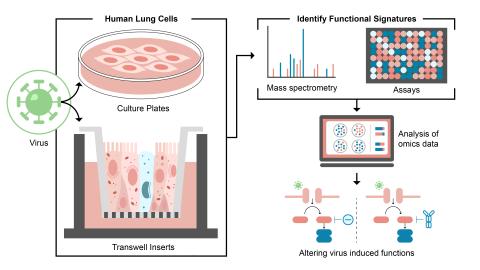
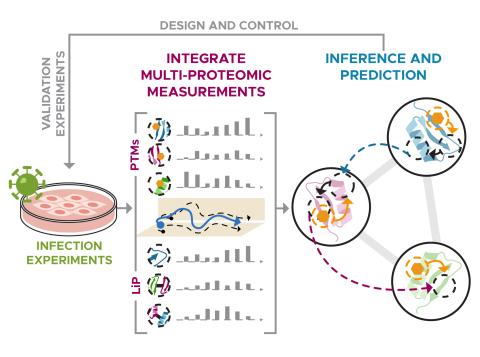
The research goal of this project is to identify and control host functions hijacked during viral infection through use of PNNL ‘omics technologies and modeling capabilities.
Datasets
0
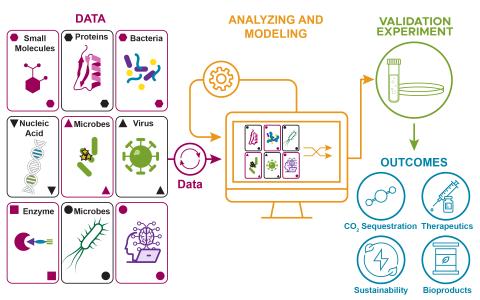
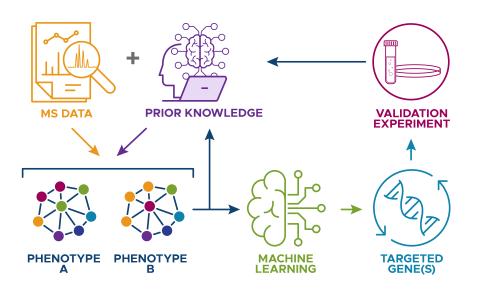
The research goal of this project is to develop a biologically informed machine learning (ML) model that integrates datasets from different studies, and leverages current biological knowledge in an automated manner, to improve predictions in biological data analysis.
Category
Datasets
0
We are constructing a streamlined approach to identify phenotype-relevant signatures by integrating various proteomics data. Leveraging protein structures and interaction networks, we will map structural changes and post-translational modifications to identify molecular drivers and subsequently...
Category
Datasets
0
By developing explainable, predictive metabolic models of individual microbes, we aim to design consortia that convert light and abundant atmospheric gases into high-value molecules through microbial division of labor.
Category
Datasets
0
The research goal of this project is to build and understand model communities that show carbon storage phenotypes
Category
Datasets
0
Samantha Powell earned her PhD from the University of Oklahoma in the lab of Dr. George Richter-Addo, using X-ray crystallography to study heme proteins and Clostridium difficile nitroreductases and their interactions with small molecules. From 2019-2020, she was a National Research Council...
Margaret S. Cheung is a biological physicist and a computational scientist on the Computing, Analytics, and Modeling team at EMSL. She graduated from the National Taiwan University in 1994 and went on to obtain a Ph.D. degree from the University of California at San Diego in 2003. She was then...
Ethan is an applied mathematician with experience in control, optimization, modeling, and machine learning. He is interested in leveraging the tools and successes of data science to push the boundaries of complexity and scale possible within scientific computing. Within modeling, his interests are...