Metagenomics is unearthing the previously hidden world of soil viruses. Many soil viral sequences in metagenomes contain putative auxiliary metabolic genes (AMGs) that are not associated with viral replication. Here, we establish that AMGs on soil viruses actually produce functional, active proteins...
Filter results
Category
- Scientific Discovery (376)
- Biology (260)
- Earth System Science (164)
- Human Health (112)
- Integrative Omics (73)
- Microbiome Science (47)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (13)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Renewable Energy (7)
- Atmospheric Science (6)
- Visual Analytics (6)
- Ecosystem Science (5)
- Coastal Science (4)
- Energy Storage (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Plant Science (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
- Water Power (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (34)
- Mass spectrometry data (32)
- Multi-Omics (32)
- Viruses (28)
- Omics (26)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- Synthetic (14)
- Genomics (13)
- sequencing (13)
- West Nile virus (13)
- High Throughput Sequencing (11)
- Influenza A (11)
- TA2 (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- TA1 (10)
- Ebola (9)
Soil viruses are highly abundant and have important roles in the regulation of host dynamics and soil ecology. Climate change is resulting in unprecedented changes to soil ecosystems and the life forms that reside there, including viruses. In this Review, we explore our current understanding of soil...
Category
The soil microbiome’s role in regulating biogeochemical processing is critical to the cycling and storage of soil organic carbon (C). The function of the microbiome under different land management uses has become a focal area of research due to the interest in managing soil C to mitigate climate...
Category
ABSTRACT Fungal mineral weathering regulates the bioavailability of inorganic nutrients from mineral surfaces to organic matter and increase the bioavailable fraction of nutrients. Such weathering strategies are classified as biomechanical or biochemical. In the case of fungal uptake of mineral...
Category
Abstract Microbial response to changing environmental factors influences the fate of soil organic carbon, and drought has been shown to affect microbial metabolism and respiration. We hypothesized that the access of microbes to different carbon pools in response to dry–rewet events occurs...
Category
Introduction: Understanding how microorganisms within a soil community interact to support collective respiration and growth remains challenging. Here, we used a model substrate, chitin, and a synthetic Model Soil Consortium (MSC-2) to investigate how individual members of a microbial community...
Category
ABSTRACT Climate change is causing an increase in drought in many soil ecosystems and a loss of soil organic carbon. Calcareous soils may partially mitigate these losses via carbon capture and storage. Here, we aimed to determine how irrigation-supplied soil moisture and perennial plants impact...
Category
Biogeochemical models for predicting carbon dynamics increasingly include microbial processes, reflecting the importance of microorganisms in regulating the movement of carbon between soils and the atmosphere. Soil viruses can redirect carbon among various chemical pools, indicating a need for...
Primary publication pre-print. Cite as: arXiv:2410.09346 [q-bio.MN]
Category
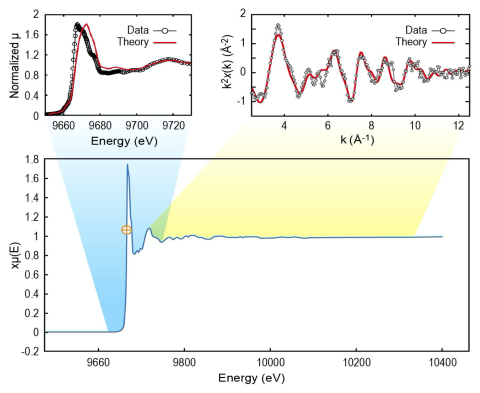
Extended X-ray absorption fine structure (EXAFS) spectroscopy is crucial for determining the coordination environment of impurities and dopants; however, it requires difficult measurements. X-ray absorption near edge structure (XANES) spectroscopy and X-ray emission spectroscopy (XES) can be...
Datasets
0
Citation: Mejia-Rodriguez D, Kim H, Sadler N, Li X, Bohutskyi P, Valiev M, Qian WJ, Cheung MS. PTM-Psi: A python package to facilitate the computational investigation of post-translational modification on protein structures and their impacts on dynamics and functions. Protein Sci. 2023 Dec;32(12)...
Category
Citation : Reichart NJ, Steiger AK, Van Fossen EM, McClure R, Overkleeft HS, Wright AT. Selection and enrichment of microbial species with an increased lignocellulolytic phenotype from a native soil microbiome by activity-based probing. ISME Commun. 2023 Sep 30;3(1):106. doi: 10.1038/s43705-023...
Category
Dr. Bohutskyi’s research focus is in developing new bioprocesses addressing sustainable transformation of carbon dioxide, biomass, and wet or liquid waste, including carbon, nitrogen, phosphorus, and other critical elements into bioproducts, chemicals, and fuels. This research uses a suite of...
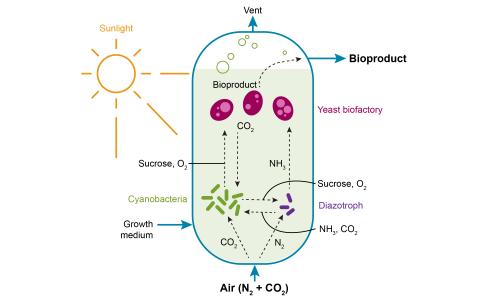
The research goal of this project is to establish model synthetic microbial communities to understand the rules regulating their biological function in order to utilize them as next generation bioproduction platforms capable of reducing carbon and nitrogen footprints in biomanufacturing processes.
Category
Datasets
4
Andrew McNaughton is a computational scientist currently working in the areas of modeling metabolic networks, multi-omics data analysis and integration, and computational chemistry. His areas of interest include high performance computing (HPC), artificial intelligence, and machine learning (ML) to...