Description
Please cite as: Naylor D.T., K.I. Naasko, M.L. Smith, S.P. Couvillion, C.D. Nicora, J.B. Trejo, and S.C. Fransen, et al. 2023. Metatranscriptomic Analysis of Soil. [Data Set] PNNL DataHub. https://doi.org/10.25584/1986545
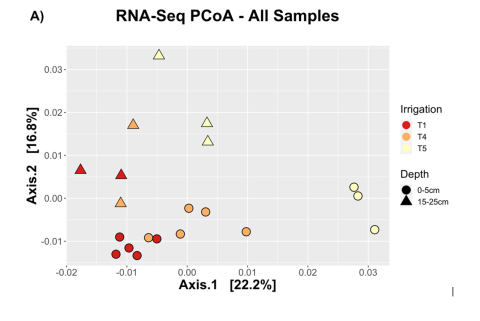
RNA was collected from soil at different depths and after three different levels of irrigation T1 100% of normal field irrigation, T4: 18.75% or normal irrigation and T5: unirrigated controls. Total RNA was isolated using the Zymo Quick-RNA fecal/soil microbe miniprep (catalog no. R2040), incorporating the DNase I treatment using Zymo’s DNase I kit (catalog no. E1010). To increase the yield of RNA, we modified the manufacturer’s instructions by first doubling the amount of soil per extraction (from 0.25 g to 0.5 g) and by performing extractions in triplicate before pooling separate extractions together. Certain soil samples (largely those from deeper soil layers) had low yield (< 100 ng per extraction) so additional rounds of extraction were performed to obtain sufficient RNA. RNA concentration was assessed using a Qubit RNA HS assay kit (Thermo Fisher) and RNA quality was determined using an Agilent 2100 BioAnalyzer (Agilent; Santa Clara, CA). The resultant RNA samples were then sequenced by GENEWIZ using Illumina technology (GENEWIZ; South Plainfield, NJ). Sequences were then aligned to a soil metagenome previously obtained from the same site using the Burrows-Wheeler aligner (BWA). SAM files were then converted to raw counts using HTSeq.