Description
Created on 2024-09-17 by LN Anderson; Last updated 2025-12-01
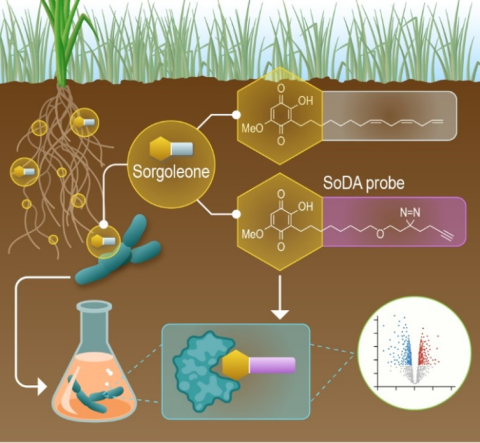
PerCon SFA: Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pitti
Mass spectrometry-based global proteome analysis and SoDA-PAL photoaffinity probe labeled protein analysis of microbial isolate Acinetobacter pittii SO1 sorgoleone interactome. DataHub download includes processed MaxQuant proteomics anlaysis (global and probe-labeled), sample naming key, updated GenBank protein locus tags (OH685), protein annotation file (.fasta), and computationally derived protein modeling (.pdb) outputs supporting data transparency and reuse.
Accessible Digital Data Downloads
Processed Proteomics and Computed Analysis
- Protein quantification by liquid chromatography mass spectrometry (LC-MS)
- AlphaFold2 protein structure predictions
Linked Primary Data Accessions
- MassIVE: MSV000091986 (global and probe-labeled proteome)
Related Sequence Collections
- GenBank: GCF_029987435.1 (genome assembly)
- RefSeq: NZ_CP125227.1 (sequence annotations)
Reference Citation(s)
- Anderson, Lindsey N, Van Fossen, Elise, Mcnaughton, Andrew D, Egbert, Robert G. SoDA-PAL Protein Profiling Proteomics of Sorgoleone Catabolizing Microbial Isolate Acinetobacter pittii SO1. United States. 2024. PNNL DataHub (Web). DOI: 10.25584/PerConSFA/2338181
- Van Fossen EM, Kroll JO, Anderson LN, McNaughton AD, Herrera D, Oda Y, Wilson AJ, Nelson WC, Kumar N, Frank AR, Elmore JR, Handakumbura P, Lin VS, Egbert RG. 0. Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pittii. Appl Environ Microbiol 0:e01026-24. DOI: 10.1128/aem.01026-24
Funding Acknowledgments
This research was supported in part by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research (BER), as part of the Genomic Science Program Secure Biosystems Design Science Focus Area Project: Persistence Control of Engineered Functions in Complex Soil Microbiomes (PerCon SFA) as a contribution of Pacific Northwest National Laboratory (PNNL). PNNL is a multiprogram national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RL01830. A portion of this research was supported under the EMSL user project award 10.46936/cpcy.proj.2022.60649/60008751, in leveraging instrument capabilities operating at the Environmental Molecular Sciences Laboratory, a DOE Office of Science User Facility, sponsored by the Biological and Environmental Research program under Contract No. DE-AC05-76RL01830.
Citation Policy
In efforts to enable discovery and reproducibility of PerCon SFA project-funded digital data assets listed at the PNNL DataHub institutional repository, we kindly ask any reuse of project data download materials acknowledge all primary and secondary dataset citations and corresponding journal articles where applicable in accordance with community best practices (e.g., FORCE11 Data Citation Principles). Reference citations, should provide the necessary metadata information required to support, corroborate, verify, and otherwise determine the legitimacy of the research findings provided (data and code) from scholarly publications and corresponding project data releases.
Data Licensing