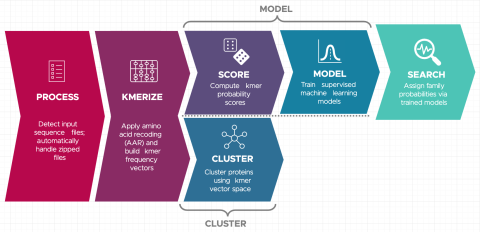
Christine H Chang, William C Nelson, Abby Jerger, Aaron T Wright, Robert G Egbert, Jason E McDermott, Snekmer: a scalable pipeline for protein sequence fingerprinting based on amino acid recoding, Bioinformatics Advances, Volume 3, Issue 1, 2023, vbad005, https://doi.org/10.1093/bioadv/vbad005.
Funding Acknowledgments
This work was supported by the Department of Energy (DOE) Office of Biological and Environmental Research (BER) and is a contribution of the Scientific Focus Area ‘Persistence Control of Engineered Functions in Complex Soil Microbiomes’, the DOE/BER ‘Improved Protein Annotation in KBase Using Machine Learning, Multi-Omics Data Integration, and Structural Prediction’ project, the DOE/BER ‘Machine-Learning Approaches for Integrating Multi-Omics Data to Expand Microbiome Annotation’ project, NSF [1856556], the Defense Threat Reduction Agency, under Interagency Agreement [DTRA13081–37739], and the Proxy Applications for Converged Workloads (PACER) project, a component of the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory. PNNL is operated for the DOE by Battelle Memorial Institute under Contract [DE-AC05-76RL01830].
Projects (1)
Last updated on 2023-02-23T19:37:46+00:00 by LN Anderson PerCon SFA Project Publication Experimental Data Catalog The Persistence Control of Engineered Functions in Complex Soil Microbiomes Project (PerCon SFA) at Pacific Northwest National Laboratory ( PNNL ) is a Genomic Sciences Program...
Datasets
5