Category
Description
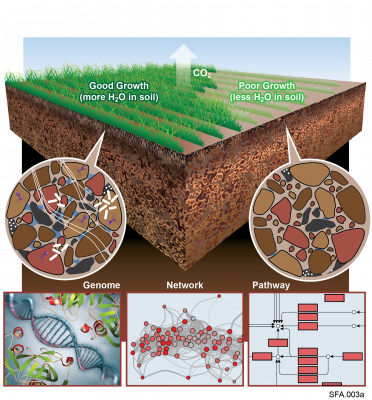
The following R source code was used for plotting figures of the viral communities detected from three grasslands soil metagenomes with a historical precipitation gradient (WA-TmG.2.0, KS-TmG.2.0, IA-TmG.2.0) from project publication 'DNA viral diversity, abundance and functional potential vary across grassland soils with a range of historical moisture regimes' resulting project data packages.
Grassland Virome Data Analysis Repository
This GitHub Repository contains viral communities detected from three grasslands with a historical precipitation gradient (IA > KS > WA).
- R codes used for plotting the figures of manuscript titled 'DNA viral diversity, abundance and functional potential vary across grassland soils with a range of historical moisture regimes' (currently submitted for review)
- R code of structure equation modeling used in the manuscript
- lysozyme hidden markov models
- viral contigs screened from the de novo assemblies of the metagenomes
Related Resource Files
Projects (1)
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
45