"DNA Viral Diversity, Abundance, and Functional Potential Vary across Grassland Soils with a Range of Historical Moisture Regimes"
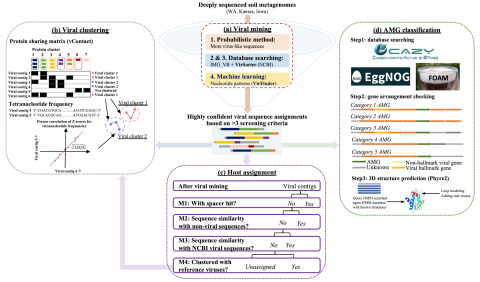
Soil viruses are abundant, but the influence of the environment and climate on soil viruses remains poorly understood. Here, we addressed this gap by comparing the diversity, abundance, lifestyle, and metabolic potential of DNA viruses in three grassland soils with historical differences in average annual precipitation, low in eastern Washington (WA), high in Iowa (IA), and intermediate in Kansas (KS). Bioinformatics analyses were applied to identify a total of 2,631 viral contigs, including 14 complete viral genomes from three deep metagenomes (1 terabase [Tb] each) that were sequenced from bulk soil DNA. An additional three replicate metagenomes (∼0.5 Tb each) were obtained from each location for statistical comparisons. Identified viruses were primarily bacteriophages targeting dominant bacterial taxa. Both viral and host diversity were higher in soil with lower precipitation. Viral abundance was also significantly higher in the arid WA location than in IA and KS. More lysogenic markers and fewer clustered regularly interspaced short palindromic repeats (CRISPR) spacer hits were found in WA, reflecting more lysogeny in historically drier soil. More putative auxiliary metabolic genes (AMGs) were also detected in WA than in the historically wetter locations. The AMGs occurring in 18 pathways could potentially contribute to carbon metabolism and energy acquisition in their hosts. Structural equation modeling (SEM) suggested that historical precipitation influenced viral life cycle and selection of AMGs. The observed and predicted relationships between soil viruses and various biotic and abiotic variables have value for predicting viral responses to environmental change.
Terabase soil metagenome samples were collected from Washington State University’s Irrigated Agriculture Research and Extension Center (IAREC), Konza Prairie Biological Station (KPBS), and Iowa State University’s Comparison of Biofuel Systems (COBS). Soil samples were collected from the same independent soil site collection tubes as the first Terabase Metagenome release, however were not pooled prior to sequencing as referenced in versions 1.0 for all three sites. The versions described in this paper are the second version (2.0), for which dataset downloads contain raw read sequencing files, assembly files, functional annotations, MIMS.me.soil.5.0 metadata information, and a dataset download “Read Me” file.
KEYWORDS: lysogeny, auxiliary metabolic gene, grassland soil, metagenome, soil bacteriophage, soil virus
Wu R, Davison MR, Nelson WC, Graham EB, Fansler SJ, Farris Y, Bell SL, Godinez I, Mcdermott JE, Hofmockel KS, Jansson JK. DNA Viral Diversity, Abundance, and Functional Potential Vary across Grassland Soils with a Range of Historical Moisture Regimes. mBio. 2021 Nov 2;12(6):e0259521. 10.1128/mBio.02595-21
Projects (1)
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
45