Filter results
Category
- Scientific Discovery (79)
- Biology (56)
- Earth System Science (26)
- Human Health (23)
- Integrative Omics (20)
- Microbiome Science (18)
- Computational Research (12)
- Chemistry (8)
- National Security (7)
- Data Analytics & Machine Learning (6)
- Computing & Analytics (5)
- Computational Mathematics & Statistics (3)
- Energy Resiliency (3)
- Chemical & Biological Signatures Science (2)
- Ecosystem Science (2)
- Materials Science (2)
- Renewable Energy (2)
- Weapons of Mass Effect (2)
- Coastal Science (1)
- Data Analytics & Machine Learning (1)
- Energy Efficiency (1)
- Energy Storage (1)
- Plant Science (1)
- Solar Energy (1)
Institution Type
Tags
- Omics (17)
- Mass Spectrometry (8)
- Virology (8)
- Multi-Omics (7)
- Genomics (6)
- High Throughput Sequencing (6)
- Mass spectrometry data (6)
- Omics-LHV Project (6)
- Proteomics (6)
- Differential Expression Analysis (5)
- Gene expression profile data (5)
- Imaging (5)
- Immune Response (5)
- Mass Spectrometer (5)
- Sequencer System (5)
- Time Sampled Measurement Datasets (5)
- Autoimmunity (4)
- Biomarkers (4)
- Homo sapiens (4)
- Molecular Profiling (4)
- Synthetic (4)
- Type 1 Diabetes (4)
- Machine Learning (3)
- Mass spectrometry-based Omics (3)
- metabolomics (3)
- Microscopy (3)
- Mus musculus (3)
- Spectroscopy (3)
- Synthetic Biology (3)
- Spectrographic Analysis (2)
Category
Dr. Nelson’s background is in genomic analysis of microbial organisms. He spent 10 years at The Institute for Genomic Research (TIGR), first as a data analyst and then as the team lead for Microbial Annotation. He was involved in some of the earliest bacterial genome sequencing projects, including...
Earth Scientist at PNNL Disciplines and Skills: Biogeochemical Cycles Carbon Cycle Isotopes Microbial Communities Microbial Ecology Microbiology Microbiome Science Nitrogen Education: Morningside College : Bachelor of Science, Biology
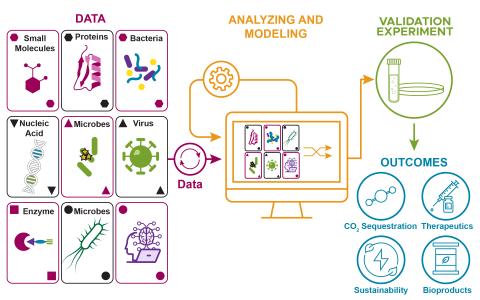
The research goal of this project is to develop computational methods to predict cell regulation phenotypes using small molecule and proteome data to understand outcomes in complex biological systems.
Datasets
0
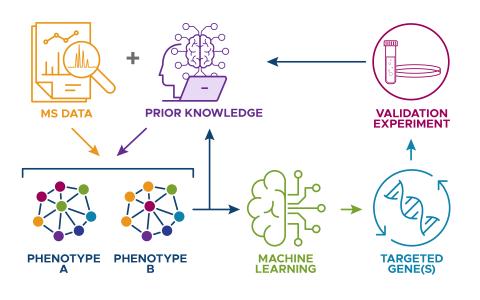
The research goal of this project is to develop a biologically informed machine learning (ML) model that integrates datasets from different studies, and leverages current biological knowledge in an automated manner, to improve predictions in biological data analysis.
Datasets
0
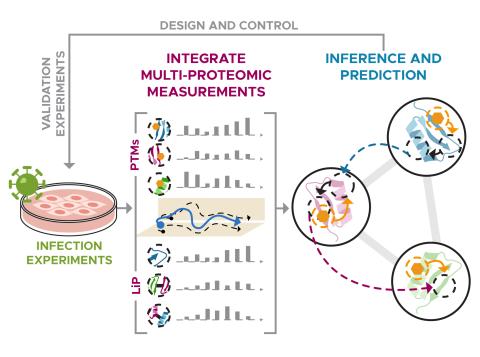
We are constructing a streamlined approach to identify phenotype-relevant signatures by integrating various proteomics data. Leveraging protein structures and interaction networks, we will map structural changes and post-translational modifications to identify molecular drivers and subsequently...
Datasets
0
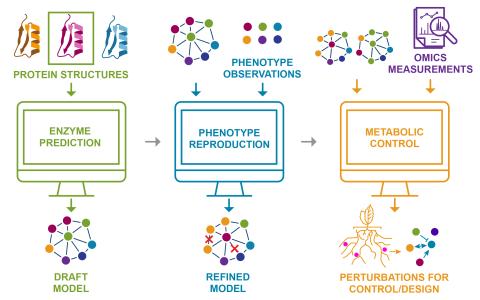
By developing explainable, predictive metabolic models of individual microbes, we aim to design consortia that convert light and abundant atmospheric gases into high-value molecules through microbial division of labor.
Datasets
0
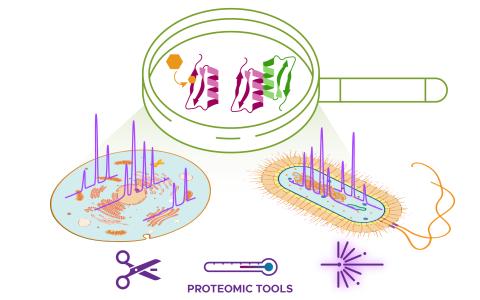
The science objective of this project is to apply structural proteomics technologies to map the molecular interactome.
Datasets
0
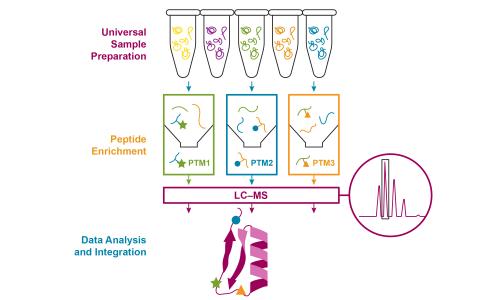
The research objective of this project is to develop an integrative and automated multi-PTM profiling capability with deep proteome coverage.
Datasets
0
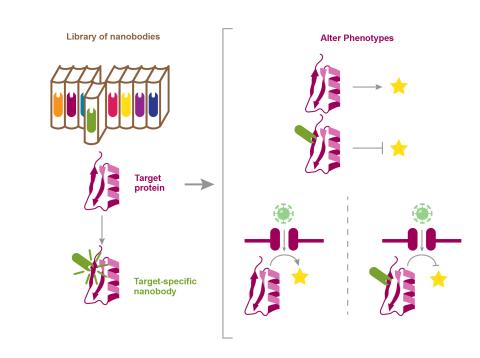
The research goal of this project is to use stimuli-specific, synthetic nanobodies to target functional mediators without prior knowledge of the response networks or manipulating the biological system.
Datasets
0
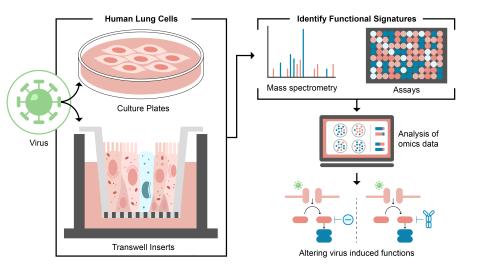
The research goal of this project is to identify and control host functions hijacked during viral infection through use of PNNL ‘omics technologies and modeling capabilities.
Datasets
0