Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL002 The purpose of this experiment was to evaluate the human host response to wild-type Middle Eastern Respiratory Syndrome coronavirus (MERS-CoV) and mutants icMERS-RFP, icMERS-DNSP16, and icMERS-d4B virus infection...
Filter results
Category
- (-) Biology (287)
- Scientific Discovery (406)
- Earth System Science (169)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (16)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (27)
- Omics (25)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Microbiome (8)
- Post-Translational Modifications (8)
- Microarray (7)
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL001 The purpose of this experiment was to evaluate the human host response to wild-type Middle Eastern Respiratory Syndrome coronavirus (MERS-CoV), and mutants icMERS-CoV-RFP, icMERS-CoV-dNSP16, icMERS-CoV-d4B, and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Interferon Experiment IFNaHUH001 The purpose of this experiment was to evaluate the human host cellular response to treatment with and without interferon alpha/beta (IFNα/β) treatment. Sample time course data was obtained from human hepatoma...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Interferon Experiment IFNMVE001 The purpose of this experiment was to evaluate the human host interferon-stimulated cellular response to interferon alpha/beta (IFNαβ) or gamma (INFγ) treatment. Sample data was obtained from primary lung...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Interferon Experiment IFNFB001 The purpose of this experiment was to evaluate the human host cellular response to interferon alpha/beta (IFNα/β) or interferon gamma (IFNγ) treatment. Sample data was obtained from primary human lung fibroblast...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Interferon Experiment IFNaIHH001 The purpose of this experiment was to evaluate the host interferon-stimulated cellular response to interferon alpha (IFNα) treatment. Sample data was obtained from human immortalized human hepatocyte cells (IHH...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Interferon Experiment IFNaCL001 The purpose of this experiment was to evaluate the host interferon-stimulated cellular response to interferon alpha (IFNα) treatment. Sample data was obtained from human lung adenocarcinoma cells (Calu-3)...
Category
Category
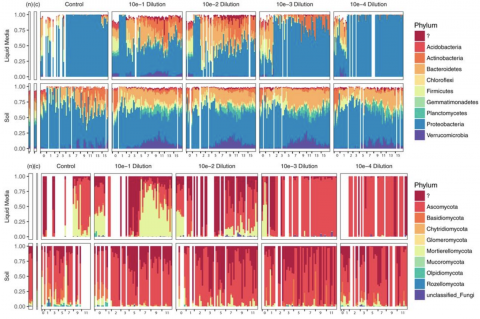
The data accompanies the manuscript that evaluates salinity-associated shifts in organic C thermodynamics, biochemical transformations, and heteroatom content in a first-order coastal watershed in the Olympic Peninsula of Washington state, USA. The files contain raw data including soil chemical...
Category
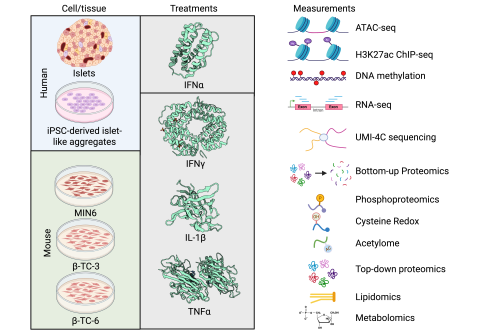
Background In type 1 diabetes (T1D), autoimmune response and inflammation cause the death of pancreatic β cells, leading to the body’s inability to produce insulin and maintain glucose homeostasis. This process is at least in part mediated by pro-inflammatory cytokines, such as interferon (IFN)α...
Category
Datasets
21
This study investigates how circadian clock regulation impacts carbon partitioning between storage, growth, and product synthesis in Synechococcus elongatus PCC 7942 in providing insights for potential strategies for enhanced bioproduction. In our analysis we explore circadian regulatory rewiring of...
Category
The ability of high-density lipoprotein (HDL) to promote cellular cholesterol efflux is a more robust predictor of cardiovascular disease protection than HDL-cholesterol levels in plasma. Previously, we found that lipidated HDL containing both apolipoprotein A-I (APOA1) and A-II (APOA2) promotes...
Category
Coronaviruses (CoV) emerge suddenly from animal reservoirs to cause novel diseases in new hosts. Discovered in 2012, the Middle East respiratory syndrome coronavirus (MERS-CoV) is endemic in camels in the Middle East and is continually causing local outbreaks and epidemics. While all three newly...
Category
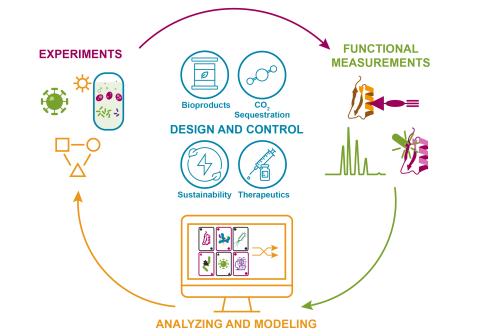
The overarching goal of this research is to predict and engineer robust microbial phenotypes under stressed conditions to accelerate and de-risk bioprocess development. Laboratory strains routinely fail to maintain productivity at industrial scales, in part due to bioprocess stresses and a limited...
Category
Datasets
0
The research goal of this project is to develop a computational approach known as Variation-leveraged Phenomic Association Study (VaLPAS) to address the challenge of using functional dark matter (proteins, metabolites, lipids) for bioeconomy applications. The project has four objectives: 1...
Category
Datasets
0