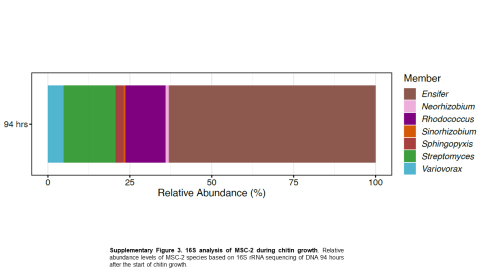
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. 16s data from MSC-2 growth. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33231 16s data from MSC-2 growth 3 fastq of 16s amplicon data of MSC2 1 csv file of raw...
Filter results
Category
- (-) Microbiome Science (50)
- (-) Energy Resiliency (13)
- Scientific Discovery (379)
- Biology (263)
- Earth System Science (166)
- Human Health (112)
- Integrative Omics (74)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Renewable Energy (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
- Water Power (1)
Tags
- Omics (10)
- PerCon SFA (9)
- High Throughput Sequencing (8)
- Genomics (7)
- Sequencer System (5)
- Synthetic Biology (5)
- Mass Spectrometry (4)
- Soil Microbiology (4)
- Amplicon Sequencing (3)
- Energy Equity (3)
- Energy Storage (3)
- A. pittii SO1 (2)
- Bioimaging (2)
- Biological and Environmental Research (2)
- Cybersecurity (2)
- Electrical energy (2)
- Heat Exchanger (2)
- Hydrothermal Liquefaction (2)
- Imaging (2)
- Long Read Sequencer (2)
- Mass spectrometry data (2)
- Proteomics (2)
- Rhizosphere Soil (2)
- RNA Sequence Analysis (2)
- Soil (2)
- Sorghum bicolor (2)
- Spectroscopy (2)
- Statistical Expression Analysis (2)
- Whole Genome Sequencing (2)
- Viruses (1)
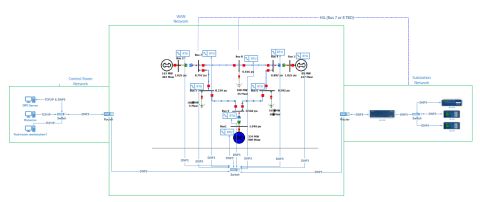
This dataset includes one baseline and three cybersecurity based scenarios utilizing the IEEE 9 Bus Model. This instantiation of the IEEE 9 model was built utilizing the OpalRT Simulator ePhasorsim module, with Bus 7 represented by hardware in the loop (HiL). The HiL was represented by two SEL351s...
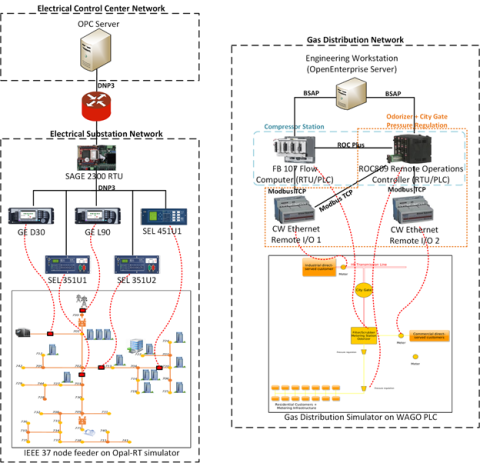
This dataset includes the results of high-fidelity, hardware in the loop experimentation on simulated models of representative electric and natural gas distribution systems with real cyber attack test cases. Such datasets are extremely important not only in understanding the system behavior during...
This data set provides ITS fungal community composition via DNA and cDNA sequence analysis at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were extracted using a Qiagen...
Category
This data set provides the ITS fungal community composition via DNA and cDNA sequence analysis for peat and sand ingrowth cores collected before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE)...
Category
This data set provides the 16S microbial community composition via DNA sequence analysis from ingrowth peat and sand cores at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the...
Category
This data set provides the peat microbial biomass carbon (MBC) and nitrogen (MBN), extractable organic carbon (EOC) and extractable nitrogen (EN) at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatland Responses Under...
Category
This data set provides the ingrowth peat microbial biomass carbon (MBC) and nitrogen (MBN), extractable organic carbon (EOC) and extractable nitrogen (EN) for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatland Responses Under Changing Environments...
Category
This data set provides the ingrowth peat extracellular enzyme potential (EE) for before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE). EE potential was quantified and calculated following a...
Category
This data set provides the 16S microbial community composition of peat and sand ingrowth cores via DNA and cDNA sequence analysis before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were...
Category
This data set provides the ingrowth peat extracellular enzyme potential (EE) for before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE). EE potential was quantified and calculated following a...
Category
This data set provides the 16S microbial community composition via DNA sequence analysis at the time of peat coring at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the Spruce and...
Category
This data set provides ITS fungal community composition via DNA sequence analysis at the time of peat coring at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the Spruce and...
Category
This data set provides the 16S microbial community composition via DNA and cDNA sequence analyses at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were extracted using a...
Category
This data set provides the ITS fungal community composition via DNA sequence analysis from sand and peat ingrowth cores at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the Spruce...