Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...
Filter results
Content type
Dataset Type
- (-) Genomics (4)
- Omics (104)
- Transcriptomics (54)
- Proteomics (50)
- Sequencing (39)
- Metabolomics (30)
- Lipidomics (28)
- Simulation Results (21)
- Amplicon (16S, ITS) (15)
- Metagenomics (9)
- Microarray (9)
- Computed Analysis (7)
- Geospacial (3)
- Microscopy (3)
- ChIP-seq (2)
- Phenomics (2)
- Staff (2)
- Cybersecurity (1)
- Electromagnetic Spectrum (1)
- Mass Spectra (1)
- Raw Image (1)
- Whole genome sequencing (WGS) (1)
Showing 1 - 4 of 4
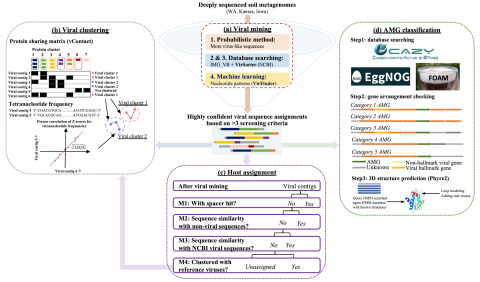
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
Dataset
Viral genome assembly annotations from terabase metagenomes (TmG.1.0) from uncultivated soil collections from the IAREC field sites in Washington (IAREC), Kansas (KBPS), and Iowa (COBS), USA. Uncultivated virus Genomes (identified from soil metagenome datasets) Data DOI Package.
Category
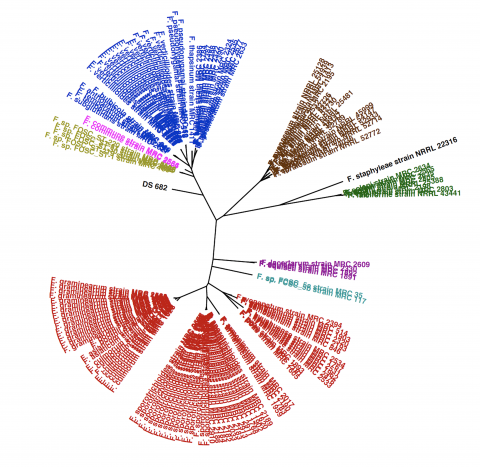
Please cite as : Bhattacharjee A., L.N. Anderson, T.D. Alfaro, A. Porras-Alfarro, A. Jumpponen, K.S. Hofmockel, and J.K. Jansson, et al. 2020. KS4A-IsoG.1.0_FspDS682 (Fungal Monoisolate Genome, KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/KS4AIsoGFspDS682/1635527 The novel fungal strain...