Data Sample
Category
Description
Please cite as: Bhattacharjee A., L.N. Anderson, T.D. Alfaro, A. Porras-Alfarro, A. Jumpponen, K.S. Hofmockel, and J.K. Jansson, et al. 2020. KS4A-IsoG.1.0_FspDS682 (Fungal Monoisolate Genome, KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/KS4AIsoGFspDS682/1635527
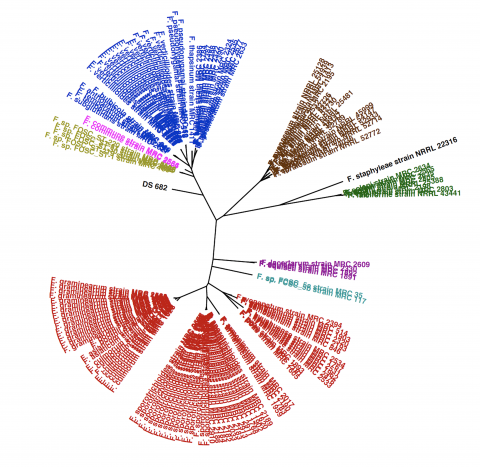
The novel fungal strain, Fusarium sp. DS 682, was isolated from the rhizosphere of the perennial grass, Bouteloua gracilis, at the Konza Prairie Biological Station in Kansas. This fungal strain is common across North American grasslands and is resilient to environmental fluctuations. The draft genome is estimated to be 97.2% complete.
Genomic sequencing of one or more organisms, or genomes in general, such as meta-information on genomes, genome projects, gene names of a given organism within a natural environment. The version described in this paper is the first version. Dataset download contains raw read sequencing files, assembly files, functional annotations, MIGS.eu.soil.5.0 metadata information, and a dataset “Read Me” file.
Data Package Resource Files:
- ReadMe_KS4A-IsoG.1.0_FspDS682.pdf (package metadata)
- MIGS.eu.soil.5.0_KS4A-IsoG.1.0_FspDS682.xlsx (sample metadata)
Items below available from Download Button:
- KS4A-IsoG.1.0_FspDS682 Functional Annotations [7.5 GB; 2 items]
- KS4A-IsoG.1.0_FspDS682 Assembly [55.9 MB; 2 items]
- KS4A-IsoG.1.0_FspDS682 Raw Reads [31.8 GB; 2 items]
Data Relationships
Parent Metagenome: KS-TmG.1.0
Omics Dataset: KS4A-Omics1.0_FspDS682
Experimental Data Accessions
- BioProject: PRJNA664411
- BioSample: SAMN16214050
- Taxonomy: 2773459
- GenBank: JACYFE000000000
- Assembly: GCA_014764975.1
- WGS: JACYFE010000001-JACYFE010011597