Category
Description
Last updated on 2025-01-16T22:12:33+00:00 by LN Anderson
Fungal Monoisolate Multi-Omics Data Package DOI "KS4A-Omics1.0_FspDS68"
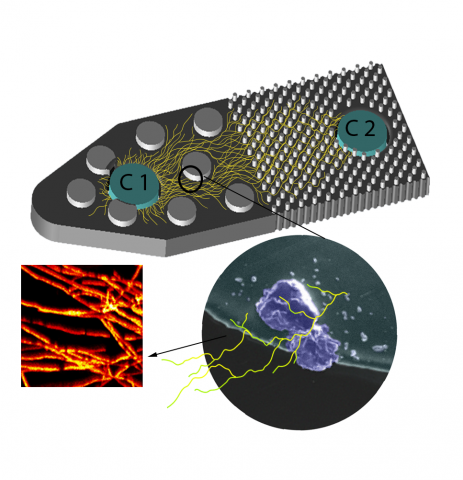
Molecular mechanisms underlying fungal mineral weathering and nutrient translocation in low nutrient environments remain poorly resolved, due to the lack of a platform for spatial analysis of biotic weathering processes. The publication data provided below, serves to address this knowledge gap by developing a mineral doped soil micromodel platform utilizing novel fungal isolate Fusarium sp. DS 682. By demonstrating the function of this platform by directly probing fungal growth using spatially resolved optical and chemical imaging methodologies, we were able to provide new evidence and visualization into fungal hyphal transport of mineral derived nutrients under nutrient and water stresses.
Data package contents reported here are the first version and contain both primary and secondary dataset deliverables, along with subsequent downstream analysis files. Data source instrument method techniques for Mass Spectroscopy, Mass Spectrometry, Microscopy, and Optimal Imaging EMSL capabilities. This data package DOI is a comprehensive collection of high-throughput multi-omics data and process method metadata. Reported data download contents are structured for compliance with reported guidelines provided by community standard initiatives and publisher stakeholder policies supporting FAIR data principles.
Accessible Digital Data DOI Downloads
Support files include additional data download “Read Me” inclusive of dataset descriptor information and source method application ontologies. Statistical software data processing workflow and resulting interactive trelliscope plots can be found at the Fusarium sp. DS682 Proteogenomics GitHub repository.
Size: 16.62 GB for 156 items
Data Types [file formats]
- Time-of-Flight Secondary Ion Mass Spectroscopy (ToF-SIMS) [ita, itm, jpg]
- Liquid Chromatography Mass Spectrometry (LC-MS) [xlsx, Thermo .RAW, mzML]
- X-Ray Diffraction (XRD) [asc, xrdml]
- X-Ray Photoelectron Spectroscopy (XPS) [csv, docx]
- X-Ray Absorption Near Edge Structure Spectroscopy (XANES) [dat, nor/avg.nor, xlsx]
- Micro-X-Ray Fluorescence Microscopy (XRF) [hdf5]
- Scanning Electron Microscopy (SEM) [tiff, pdf]
- Optical Microscopy (OM) [tiff, jpg]
Processed Files
- Fusarium sp. DS 682 Protein Sequence Annotations [GenBank]
- Comprehensive Proteogenomic Annotations & KEGG Mapping
- Statistical Proteomics Analysis & Interactive Trelliscope Plots
Linked Open Primary Data
Primary mass spectrometry proteome and corresponding parameter files (xml) have been deposited at the Mass Spectrometry Interactive Virtual Environment (MassIVE), public domain community data repository, promoting the free exchange of mass spectrometry data.
- MassIVE Accession: MSV000087221
- MassIVE DOI: 10.25345/C50V4N
Related Software Tools
- SMAK Microprobe Analysis Toolkit
- SIXPACK
- Athena Demeter Processing Suite
- Bruker TOPAZ
- Oxford INCA
- MultiPak
- ImageJ
- MaxQuant (v.1.6.7.0)
- pmartR (GitHub)
- Trelliscope (GitHub)
- iPath3
- SurfaceLab (v.6.4)
Acknowledgment of Federal Funding
This research was supported by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research (BER) and is a contribution of the Scientific Focus Area "Phenotypic response of the soil microbiome to environmental perturbations." Pacific Northwest National Laboratory (PNNL) is operated for the DOE by Battelle Memorial Institute under Contract DE-AC05-76RLO1830. A portion of the research was performed using the Environmental Molecular Sciences Laboratory, a DOE Office of Science User Facility sponsored by the BER and located at PNNL.
Reference Citations
- Anderson, Lindsey N, Bhattacharjee, Arunima, Richardson, Jocelyn R, Bramer, Lisa M, Qafoku, Odeta, Zhu, Zihua, Bowden, Mark E, Engelhard, Mark H, Nelson, William C, Hofmockel, Kirsten S, Jansson, Janet K, Anderton, Christopher R. 2021. KS4A-Omics1.0_FspDS682. [Data Set] PNNL DataHub. https://doi.org/10.25584/KSOmicsFspDS682/1766303.
- Bhattacharjee A, Qafoku O, Richardson JA, Anderson LN, Schwarz K, Bramer LM, Lomas GX, Orton DJ, Zhu Z, Engelhard MH, Bowden ME, Nelson WC, Jumpponen A, Jansson JK, Hofmockel KS, Anderton CR. (2022). A Mineral-Doped Micromodel Platform Demonstrates Fungal Bridging of Carbon Hot Spots and Hyphal Transport of Mineral-Derived Nutrients. mSystems, e0091322. Advance online publication. https://doi.org/10.1128/msystems.00913-22.