The National Park Air Quality Index dataset (NPS-AQI) consists of spatially and temporally matched images and air quality measurements from 16 different national parks. The images are taken from the National Park Service's publicly available air quality web cameras, and the air pollutant and...
Filter results
Category
- Scientific Discovery (369)
- Biology (258)
- Earth System Science (161)
- Human Health (112)
- Integrative Omics (73)
- Microbiome Science (47)
- National Security (31)
- Computational Research (25)
- Computing & Analytics (17)
- Chemical & Biological Signatures Science (12)
- Energy Resiliency (12)
- Weapons of Mass Effect (12)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Materials Science (7)
- Atmospheric Science (6)
- Data Analytics & Machine Learning (6)
- Renewable Energy (6)
- Visual Analytics (6)
- Coastal Science (4)
- Ecosystem Science (4)
- Energy Storage (3)
- Plant Science (3)
- Solar Energy (3)
- Bioenergy Technologies (2)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Grid Cybersecurity (2)
- Transportation (2)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Wind Energy (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (34)
- Mass spectrometry data (32)
- Multi-Omics (32)
- Viruses (28)
- Omics (26)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- Synthetic (14)
- Genomics (13)
- sequencing (13)
- West Nile virus (13)
- High Throughput Sequencing (11)
- Influenza A (11)
- TA2 (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- TA1 (10)
- Ebola (9)
This dataset consists of input files and model output from a large eddy simulation of clouds. It is based on conditions observed during the Cloud System Evolution in the Trades (CSET) field campaign on 7 Aug. 2015, using a baseline aerosol concentration estimated from measurements. It is a single...
This dataset consists of input files and model output from a large eddy simulation of clouds. It is based on conditions observed during Aerosol and Cloud Experiments in the Eastern North Atlantic (ACE-ENA) and at the Atmospheric Radiation Measurement (ARM) program Eastern North Atlantic (ENA)...
This dataset consists of input files and model output from a large eddy simulation of clouds. It is based on conditions observed during Aerosol and Cloud Experiments in the Eastern North Atlantic (ACE-ENA) and at the Atmospheric Radiation Measurement (ARM) program Eastern North Atlantic (ENA)...
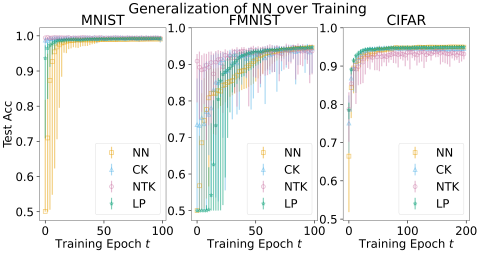
Small CNN Experiment Data Repository DataBase POC: andrew.engel@pnnl.gov This repository contains the data products created for the https://arxiv.org/pdf/2310.18612, "Efficient kernel surrogates for neural network-based regression", submitted to PNAS. As outlined in section 6.3, a small CNN model...
Ice nucleating particles (INPs) are rare particles that initiate primary ice formation, a critical step required for subsequent important cloud microphysical processes that ultimately govern cloud phase and cloud radiative properties. Laboratory studies have found that organic-rich dusts, such as...
Ice nucleating particles (INPs) are rare particles that initiate primary ice formation, a critical step required for subsequent important cloud microphysical processes that ultimately govern cloud phase and cloud radiative properties. Laboratory studies have found that organic-rich dusts, such as...
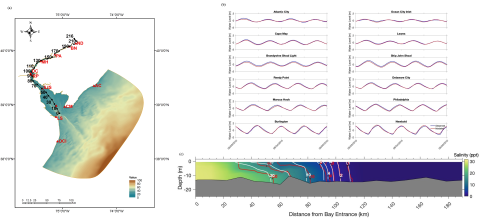
This dataset (R1_Baseline_Bot_S_RiverKilometer.nc) contains the model result from Case R1 (baseline, Sea Level Rise = 0m). All the variables in the netcdf file are obtained at the points along the River Kilometer (RK) system measuring upstream from the river mouth (RK=0), which are defined as...
Comprised of 6,426 sample runs, The Environmental Determinants of Diabetes in the Young (TEDDY) proteomics validation study constitutes one of the largest targeted proteomics studies in the literature to date. Making quality control (QC) and donor sample data available to researchers aligns with...
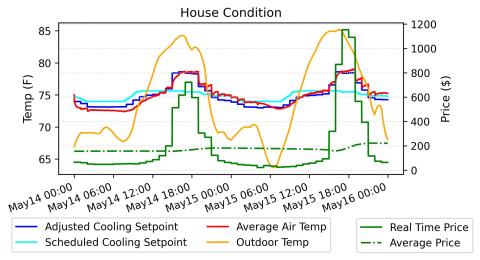
Real time pricing (RTP) is often promoted as a mechanism to improve the economic efficiency of the electricity system. However, many regulators have been hesitant to adopt RTP due to concerns about exposing customers to extreme price swings. To balance these concerns, this paper proposes a...
Federal and state decarbonization goals have led to numerous financial incentives and policies designed to increase access and adoption of renewable energy systems. In combination with the declining cost of both solar photovoltaic and battery energy storage systems and rising electric utility rates...
Category
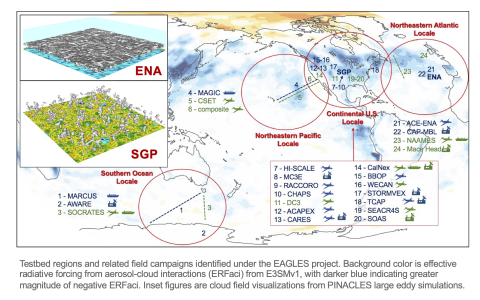
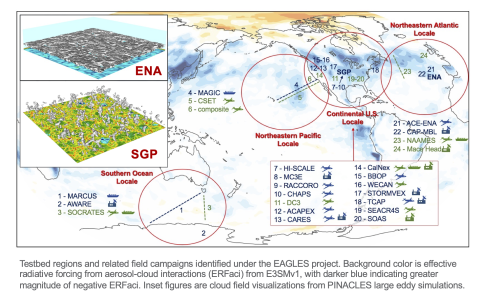
EAGLES Liquid Cloud Testbed Large Eddy Simulation Library (v2) This library consists of large eddy simulation (LES) model output using the PINACLES codebase coupled to the Hebrew University Fast Spectral Bin Microphysics scheme representing shallow, liquid phase clouds from a range of global liquid...
EAGLES Liquid Cloud Testbed Large Eddy Simulation Library (v2) This library consists of large eddy simulation (LES) model output using the PINACLES codebase coupled to the Hebrew University Fast Spectral Bin Microphysics scheme representing shallow, liquid phase clouds from a range of global liquid...
PNNL-SA-192987 This dataset contains annual mean and 6-hourly output data from a set of E3SMv1 simulations that demonstrate the impact of numerical process coupling on the simulated dust life cycle. The simulations are documented and analyzed in the attached manuscript (pdf file). Publishing the...
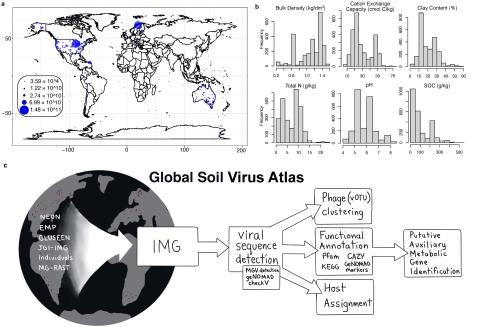
Please cite as : Graham E.B., A. Camargo, R. Wu, R.Y. Neches, M. Nolan, W.C. Nelson, and M.M. Pleake, et al. 2024. Global Soil Virus (GSV) Atlas. [Data Set] PNNL DataHub. 10.25584/2229733 This data is published under a CC0 license . The authors encourage data reuse and request attribution by...