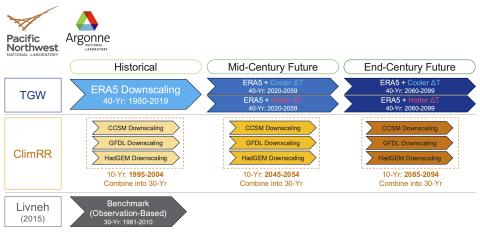
The current data upload contains the following hydrologic (water balance) simulation data over CONUS: Climate Forcing: ClimRR (CCSM) ( https://climrr.anl.gov/climrrdata ) Simulation Scenario: Historical Simulation Model: VIC (Variable Infiltration Capacity) Output Variables: OUT_EVAP Output...
Filter results
Category
- Scientific Discovery (406)
- Biology (287)
- Earth System Science (169)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (16)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (28)
- Omics (26)
- Mass Spectrometry (25)
- Soil Microbiology (25)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- Synthetic (14)
- Genomics (13)
- sequencing (13)
- West Nile virus (13)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Machine Learning (8)
- Microbiome (8)
The current data upload contains the following river routing (water management) simulation data over CONUS: Climate Forcing: ClimRR (CCSM) ( https://climrr.anl.gov/climrrdata ) Simulation Scenario: End-Century (RCP85) Simulation Model: mosartwmpy (Model for Scale Adaptive River Transport-Water...
The current dataset contains data upload links to the following river routing (water management) simulation data over CONUS: Climate Forcing: ClimRR (CCSM) ( https://climrr.anl.gov/climrrdata ) Simulation Scenario: Historical Simulation Period: 1995-2004 Simulation Model: mosartwmpy (Model for Scale...
The current data upload contains the following hydrologic (water balance) simulation data over CONUS: Climate Forcing: TGW (Cooler) ( https://tgw-data.msdlive.org/ ) Simulation Scenario: Mid-Century (RCP85) Simulation Model: VIC (Variable Infiltration Capacity) Output Variables: OUT_SOIL_WET Output...
The current data upload contains the following hydrologic (water balance) simulation data over CONUS: Climate Forcing: ClimRR (HadGEM) ( https://climrr.anl.gov/climrrdata ) Simulation Scenario: Mid-Century (RCP85) Simulation Model: VIC (Variable Infiltration Capacity) Output Variables: OUT_SWE...
The current data upload contains the following hydrologic (water balance) simulation data over CONUS: Climate Forcing: TGW (Hotter) ( https://tgw-data.msdlive.org/ ) Simulation Scenario: Mid-Century (RCP85) Simulation Model: VIC (Variable Infiltration Capacity) Output Variables: OUT_EVAP Output...
The current data upload contains the following river routing (water management) simulation data over CONUS: Climate Forcing: ClimRR (HadGEM) ( https://climrr.anl.gov/climrrdata ) Simulation Scenario: End-Century (RCP85) Simulation Model: mosartwmpy (Model for Scale Adaptive River Transport-Water...
The current data upload contains the following hydrologic (water balance) simulation data over CONUS: Climate Forcing: TGW (Cooler) ( https://tgw-data.msdlive.org/ ) Simulation Scenario: Mid-Century (RCP85) Simulation Model: VIC (Variable Infiltration Capacity) Output Variables: OUT_BASEFLOW Output...
The current data upload contains the following hydrologic (water balance) simulation data over CONUS: Climate Forcing: Livneh ( https://www.nature.com/articles/sdata201542 ) Simulation Scenario: Historical Simulation Model: VIC (Variable Infiltration Capacity) Output Variables: OUT_SWE Output...
The current data upload contains the following river routing (water management) simulation data over CONUS: Climate Forcing: TGW ( https://tgw-data.msdlive.org/ ) Simulation Scenario: Historical Simulation Model: mosartwmpy (Model for Scale Adaptive River Transport-Water Management in Python) Output...
The current data upload contains the following hydrologic (water balance) simulation data over CONUS: Climate Forcing: TGW (Cooler) ( https://tgw-data.msdlive.org/ ) Simulation Scenario: End-Century (RCP85) Simulation Model: VIC (Variable Infiltration Capacity) Output Variables: OUT_RUNOFF Output...
The current data upload contains the following hydropower simulation data: Climate Forcing: ClimRR (GFDL) ( https://climrr.anl.gov/climrrdata ) Simulation Scenario: End-Century (RCP85) Simulation Period: 2085-2094 Simulation Model: PNNL B1Hydro Output Format: CSV Spatial Resolution: Hydropower...
The current data upload contains the following river routing (water management) simulation data over CONUS: Climate Forcing: TGW (Hotter) ( https://tgw-data.msdlive.org/ ) Simulation Scenario: End-Century (RCP85) Simulation Model: mosartwmpy (Model for Scale Adaptive River Transport-Water Management...
The current data upload contains the following river routing (water management) simulation data over CONUS: Climate Forcing: ClimRR (GFDL) ( https://climrr.anl.gov/climrrdata ) Simulation Scenario: Historical Simulation Model: mosartwmpy (Model for Scale Adaptive River Transport-Water Management in...
The current data upload contains the following hydrologic (water balance) simulation data over CONUS: Climate Forcing: TGW (Hotter) ( https://tgw-data.msdlive.org/ ) Simulation Scenario: Mid-Century (RCP85) Simulation Model: VIC (Variable Infiltration Capacity) Output Variables: OUT_SWE Output...