Dataset Description The purpose of this experiment was to evaluate the regulatory stress response of Oleaginous yeast species Rhodotorula toruloides NBRC 0880 (JGI strain IFFO0880 v4.0) under nitrogen-rich and nitrogen-limited conditions over time. Time course experimental samples (24, 48, and 72...
Filter results
Category
- (-) Biology (287)
- (-) National Security (32)
- Scientific Discovery (404)
- Earth System Science (167)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (14)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (27)
- Omics (25)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Microbiome (8)
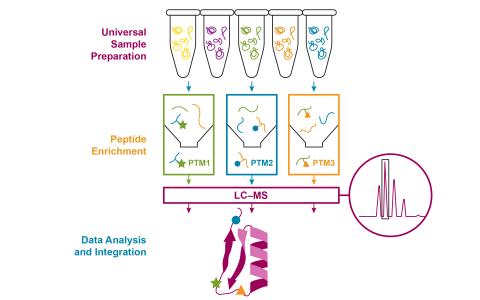
- Post-Translational Modifications (8)
- Microarray (7)
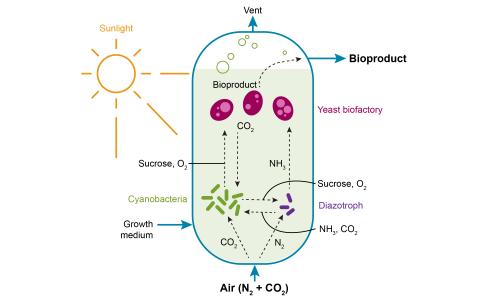
Created 2024-12-20T23:47:47+00:00 by LN Anderson ; Last updated 2025-05-09T15:55:34+00:00 S. elongatus PCC 7942 Circadian Control Bioproduction Metabolomics (PB-DP5) The purpose of this experiment was to evaluate how circadian clock regulation impacts carbon partitioning between storage, growth, and...
Category
Created 2024-12-20T23:47:47+00:00 by LN Anderson ; Last updated 2025-05-09T15:55:34+00:00 S. elongatus PCC 7942 Circadian Control Bioproduction Transcriptomics (PB-DP3) The purpose of this experiment was to evaluate how circadian clock regulation impacts carbon partitioning between storage, growth...
Category
This data has been submitted to the RSCB Protein Data Bank (PDB) under accession code 7TVO (V-Csn-E157Q).
Category
This data has been submitted to the RSCB Protein Data Bank (PDB) under accession code 7TVP (V-Csn-E157Q chitohexaose complex).
Category
Original data available on MassIVE under accession number MSV000086931 . Paper tentative title: Drought and wet-up causes significant remodeling of the soil lipidome Author list: Sneha Couvillion et al. We used lipidomics to capture the rapid metabolic response of the microbial community in an arid...
Category
This data has been submitted to the RSCB Protein Data Bank (PDB) under accession code 7TVO (V-Csn-E157Q).
Category
This data has been submitted to the RSCB Protein Data Bank (PDB) under accession code 7TVL (V-Csn apo1).
Category
This data has been deposited at the Sequence Read Archive (SRA) SRP400851 under the BioProject accession: PRJNA886747 .
Category
This data has been submitted to the RSCB Protein Data Bank (PDB) under accession code 7TVN (V-Csn-D148N).
Category
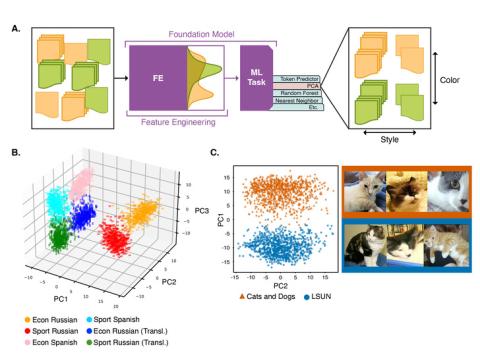
This repository contains data for the experiments run in the paper "Understanding Generative AI Content with Embedding Models" ( https://arxiv.org/abs/2408.10437 ). DataBase POC: Max Vargas (max.vargas@pnnl.gov) The data is separated by experiment: A. The `stack_exchange` dataset contains a...
Category
Please cite as : Rodríguez-Ramos, Josué, Natalie Sadler, Elias K. Zegeye, Yuliya Farris, Samuel Purvine, Sneha Couvillion, William C. Nelson, and Kirsten Hofmockel. Environmental matrix and moisture are key determinants of microbial phenotypes expressed in a reduced complexity soil-analog. 2024....
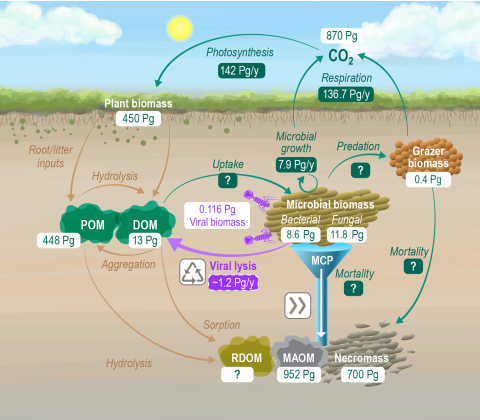
Original data published and available from Zenodo at: https://doi.org/10.5281/zenodo.13839485 Please cite as : Zimmerman, A., & Hofmockel, K. (2024). Global terrestrial carbon pools and fluxes [Data set]. In Global Change Biology. Zenodo. https://doi.org/10.5281/zenodo.13839485 This file contains a...
Category
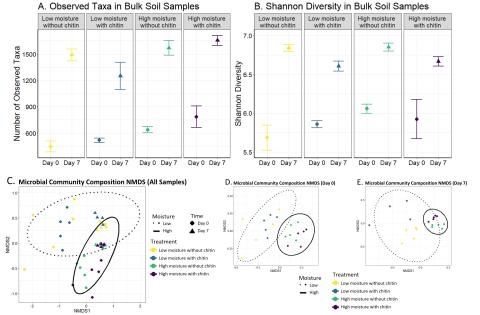
Please cite as : Nicholas J Reichart, Sheryl Bell, Vanessa A. Garayburu-Caruso, Natalie Sadler, Sharon Zhao, Kirsten Hofmockel. Chitin Decomposition 16S rRNA gene fastq. 2024 . [Data Set] PNNL DataHub. 10.25584/2475041. This data is published under a CC0 license . The authors encourage data reuse...
Category
Dataset Description The purpose of this experiment was to evaluate the human host cellular response to wild type human coronavirus strain 229E (HCoV-229E) infection. Sample data was obtained for mock-infected and HCoV-229E infected immortalized human lung epithelial cells (A549) (MOI 5) and...