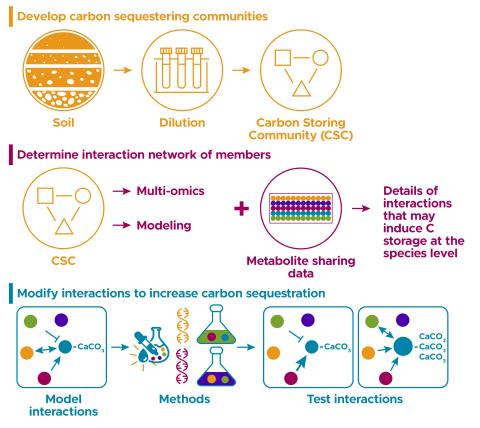
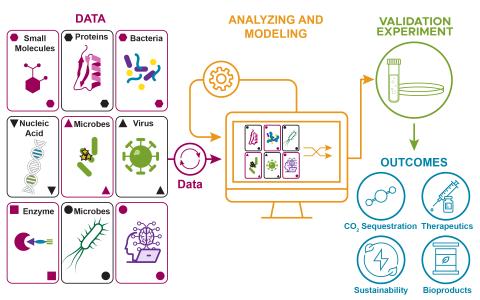
The soil microbiome performs many functions that are key to ecology, agriculture, and nutrient cycling. However, because of the complexity of this ecosystem we do not know the molecular details of the interactions between microbial species that lead to these important functions. Here, we use a...
Filter results
Category
- (-) Biology (287)
- Scientific Discovery (406)
- Earth System Science (169)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (16)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (27)
- Omics (25)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Microbiome (8)
- Post-Translational Modifications (8)
- Microarray (7)
Two factors that are well-known to influence soil microbiomes are the depth of the soil as well as the level of moisture. Previous works have demonstrated that climate change will increase the incidence of drought in soils, but it is unknown how fluctuations in moisture availability affect soil...
Category
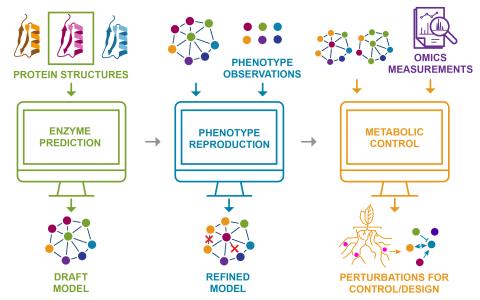
The research goal of this project is to develop new theory and tools that leverage evolutionary perspectives and knowledge of the energetics of reactions to predict the most likely regulation in a given environment. These methods will accelerate exploration, modeling and understanding of cell...
Category
Datasets
3
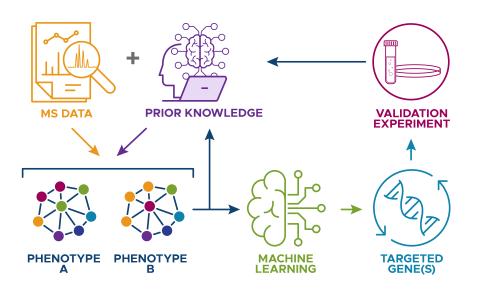
The research goal of this project is to develop a biologically informed machine learning (ML) model that integrates datasets from different studies, and leverages current biological knowledge in an automated manner, to improve predictions in biological data analysis.
Category
Datasets
0
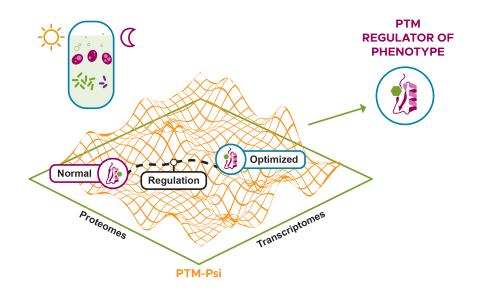
The research goal of this project is to construct and streamline an approach to identify phenotype-relevant signatures by integrating various proteomics data. Leveraging protein structures and interaction networks, we will map structural changes and post-translational modifications to identify...
Category
Datasets
0
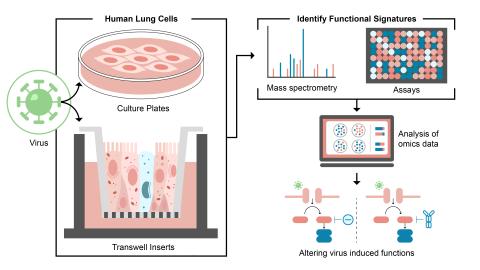
This project will improve current understanding of how viruses manipulate host environments. Use cutting edge, iterative proteomics and metabolomics tools in ex vivo primary human lung cultures that recapitulate the epithelium of the conducting airway and novel high throughput sample capture...
Datasets
5
The research goal of this project is to build and understand model communities that show carbon storage phenotypes
Category
Datasets
0
By developing explainable, predictive metabolic models of individual microbes, we aim to design consortia that convert light and abundant atmospheric gases into high-value molecules through microbial division of labor.
Category
Datasets
0
The research goal of this project is to develop computational methods to predict cell regulation phenotypes using small molecule and proteome data to understand outcomes in complex biological systems.
Category
Datasets
0
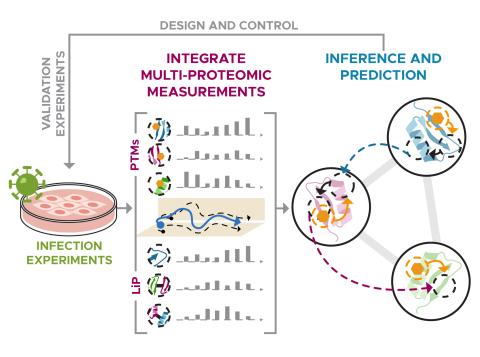
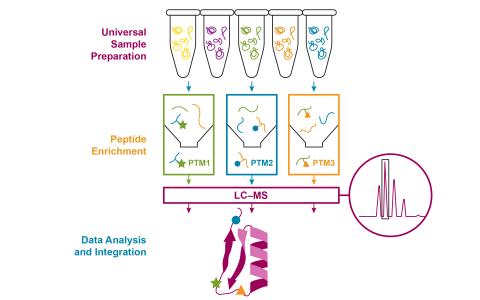
The research objective of this project is to develop an integrative and automated multi-PTM profiling capability with deep proteome coverage.
Category
Datasets
2
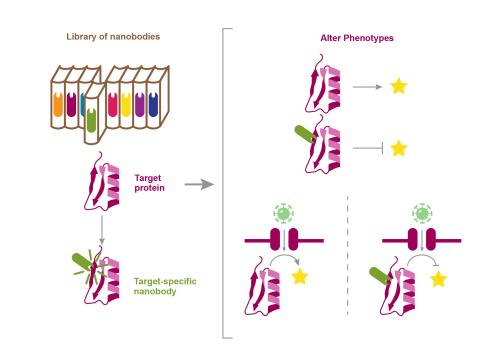
The research goal of this project is to use stimuli-specific, synthetic nanobodies to target functional mediators without prior knowledge of the response networks or manipulating the biological system.
Category
Datasets
0
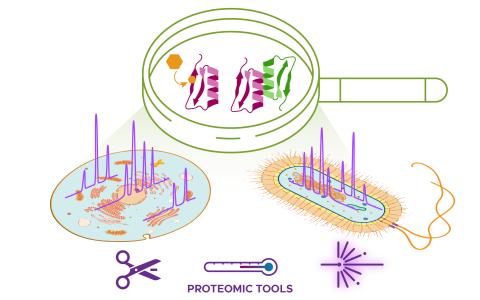
The science objective of this project is to apply structural proteomics technologies to map the molecular interactome.
Category
Datasets
5
The Human Islet Research Network (HIRN) is a large consortia with many research projects focused on understanding how beta cells are lost in type 1 diabetics (T1D) with a goal of finding how to protect against or replace the loss of functional beta cells. The consortia has multiple branches of...
Category
Datasets
1
Predictive Phenomics Initiative (PPI) Project Data Catalog Collection The Predictive Phenomics Initiative (PPI) is an internal LDRD investment at Pacific Northwest National Laboratory focused on unraveling the mysteries of molecular function in complex biological systems. Explore PPI research...
Category
Datasets
18
The science objectives of this project are to: Functionally enrich microbial communities and generate multi-omics to correlate biochemical mechanisms to activity. Integrate PhenoProfiling with Thrust Areas 2 and 3 to develop models for phenotype prediction and interspecies interactions. Evaluate...
Category
Datasets
1