Biomedical Resilience & Readiness in Adverse Operating Environments (BRAVE) Project: Exhaled Breath Condensate (EBC) TMT Proteomic Transformation Data Exhaled breath condensate (EBC) represents a low-cost and non-invasive means of examining respiratory health. EBC has been used to discover and...
Filter results
Category
- (-) Scientific Discovery (369)
- Biology (258)
- Earth System Science (161)
- Human Health (112)
- Integrative Omics (73)
- Microbiome Science (47)
- National Security (31)
- Computational Research (25)
- Computing & Analytics (17)
- Chemical & Biological Signatures Science (12)
- Energy Resiliency (12)
- Weapons of Mass Effect (12)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Materials Science (7)
- Atmospheric Science (6)
- Data Analytics & Machine Learning (6)
- Renewable Energy (6)
- Visual Analytics (6)
- Coastal Science (4)
- Ecosystem Science (4)
- Energy Storage (3)
- Plant Science (3)
- Solar Energy (3)
- Bioenergy Technologies (2)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Grid Cybersecurity (2)
- Transportation (2)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Wind Energy (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (34)
- Mass spectrometry data (32)
- Multi-Omics (32)
- Viruses (27)
- Omics (26)
- Mass Spectrometry (24)
- Health (23)
- Soil Microbiology (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- TA2 (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- TA1 (10)
- Ebola (9)
- Microbiome (8)
This data set provides the ITS fungal community composition via DNA sequence analysis from sand and peat ingrowth cores at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the Spruce...
Category
This data set provides ITS fungal community composition via DNA sequence analysis at the time of peat coring at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the Spruce and...
Category
This data set provides the peat microbial biomass carbon (MBC) and nitrogen (MBN), extractable organic carbon (EOC) and extractable nitrogen (EN) at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatland Responses Under...
Category
This data set provides the 16S microbial community composition of peat and sand ingrowth cores via DNA and cDNA sequence analysis before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were...
Category
This data set provides the ingrowth peat microbial biomass carbon (MBC) and nitrogen (MBN), extractable organic carbon (EOC) and extractable nitrogen (EN) for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatland Responses Under Changing Environments...
Category
This data set provides the 16S microbial community composition via DNA and cDNA sequence analyses at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were extracted using a...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL105 Metadata The purpose of this experiment was to evaluate the host epigenetic response to Influenza A virus (subtype H5N1) infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and...
Category
This data set provides the 16S microbial community composition via DNA sequence analysis at the time of peat coring at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the Spruce and...
Category
This data set provides the ingrowth peat extracellular enzyme potential (EE) for before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE). EE potential was quantified and calculated following a...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL106 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to Influenza A virus (subtype H5N1) infection. Samples were obtained from human lung adenocarcinoma cell line (Calu...
Category
This data set provides the ingrowth peat extracellular enzyme potential (EE) for before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE). EE potential was quantified and calculated following a...
Category
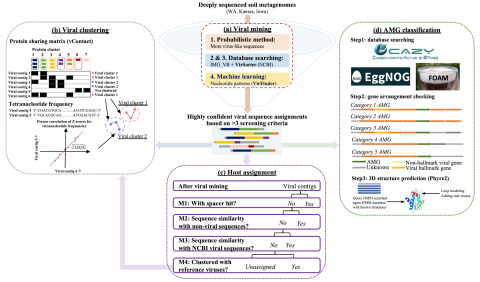
Viral genome assembly annotations from terabase metagenomes (TmG.1.0) from uncultivated soil collections from the IAREC field sites in Washington (IAREC), Kansas (KBPS), and Iowa (COBS), USA. Uncultivated virus Genomes (identified from soil metagenome datasets) Data DOI Package.
Category
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from KPBS field site in Manhattan, KS. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from IAREC field site in Prosser, WA. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.