This data set provides the ingrowth peat extracellular enzyme potential (EE) for before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE). EE potential was quantified and calculated following a...
Filter results
Category
- Scientific Discovery (178)
- Biology (106)
- Earth System Science (89)
- Human Health (69)
- Integrative Omics (52)
- Microbiome Science (16)
- National Security (14)
- Computing & Analytics (10)
- Energy Resiliency (7)
- Computational Research (6)
- Visual Analytics (6)
- Atmospheric Science (3)
- Coastal Science (3)
- Computational Mathematics & Statistics (3)
- Data Analytics & Machine Learning (3)
- Data Analytics & Machine Learning (3)
- Renewable Energy (3)
- Chemical & Biological Signatures Science (2)
- Cybersecurity (2)
- Distribution (2)
- Ecosystem Science (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Weapons of Mass Effect (2)
- Bioenergy Technologies (1)
- Chemistry (1)
- Computational Mathematics & Statistics (1)
- Energy Efficiency (1)
- Energy Storage (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Plant Science (1)
- Solar Energy (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Dataset Type
- Omics (86)
- Transcriptomics (51)
- Proteomics (39)
- Sequencing (39)
- Metabolomics (28)
- Lipidomics (26)
- Simulation Results (17)
- Amplicon (16S, ITS) (15)
- Metagenomics (9)
- Microarray (9)
- Genomics (4)
- Computed Analysis (3)
- ChIP-seq (2)
- Geospacial (2)
- Staff (2)
- Cybersecurity (1)
- Electromagnetic Spectrum (1)
- Mass Spectra (1)
- Microscopy (1)
- Phenomics (1)
- Whole genome sequencing (WGS) (1)
Tags
- Virology (63)
- Immune Response (48)
- Time Sampled Measurement Datasets (45)
- Gene expression profile data (42)
- Differential Expression Analysis (41)
- Homo sapiens (30)
- Mass spectrometry data (25)
- Multi-Omics (23)
- MERS-CoV (16)
- Mus musculus (15)
- Viruses (14)
- Health (13)
- Soil Microbiology (13)
- Virus (13)
- West Nile virus (11)
- Synthetic (10)
- Ebola (9)
- Influenza A (9)
- sequencing (9)
- Resource Metadata (8)
- Metagenomics (7)
- Microarray (5)
- Genomics (4)
- Human Interferon (4)
- Microbiome (4)
- Omics (4)
- IAREC (3)
- metagenomics (3)
- Sequencing (3)
- soil microbiology (3)
This data set provides the 16S microbial community composition via DNA and cDNA sequence analyses at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were extracted using a...
Category
This data set provides the 16S microbial community composition of peat and sand ingrowth cores via DNA and cDNA sequence analysis before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were...
Category
This data set provides ITS fungal community composition via DNA and cDNA sequence analysis at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were extracted using a Qiagen...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL106 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to Influenza A virus (subtype H5N1) infection. Samples were obtained from human lung adenocarcinoma cell line (Calu...
Category
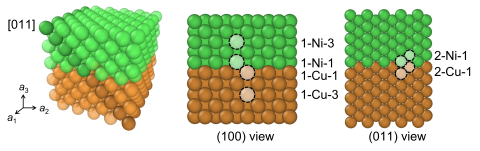
A trained neural net potential (NNP) was use to simulate shear in the [011] direction in Cu-Ni multilayers. We applied the NNP to a Monte Carlo scheme to generate parallel shear simulations, demonstrating the range of possible trajectories that can be obtained due to speedups provided by the NNP...
Simulated x-ray absorption spectra calculated with the FEFF code for various models of Cu incorporated into the iron oxide mineral hematite. The structures are drawn from publically available ab initio molecular dynamics simulations.
The PNNL-PFAS database was constructed to provide quantitative infrared spectra of per- and polyfluoroalkyl substances (PFAS) in the gas phase. These fluorine-rich, organic compounds have been used as non-flammable solvents, cleaning agents, and in the manufacture of a diverse variety of consumer...
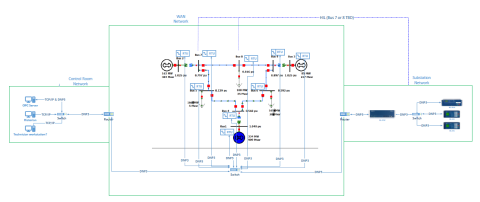
This dataset includes one baseline and three cybersecurity based scenarios utilizing the IEEE 9 Bus Model. This instantiation of the IEEE 9 model was built utilizing the OpalRT Simulator ePhasorsim module, with Bus 7 represented by hardware in the loop (HiL). The HiL was represented by two SEL351s...
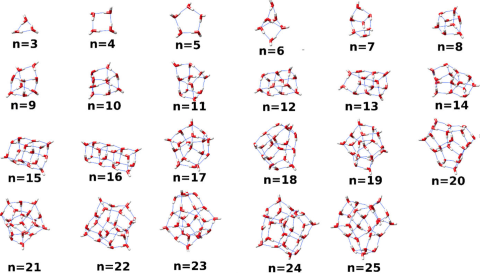
Structures of putative minima and low-energy networks for water clusters of sizes n = 3–30 as described in the manuscript by Avijit Rakshit and Pradipta Bandyopadhyay, Joseph P. Heindel and Sotiris S. Xantheas “ Atlas of putative minima and low-lying energy networks of water clusters n=3-25 ”...
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes. [Data Set] PNNL DataHub. https://doi.org/10.25584/PNNLDH/1986536 Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes...
Category
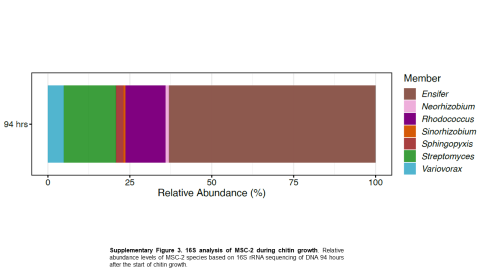
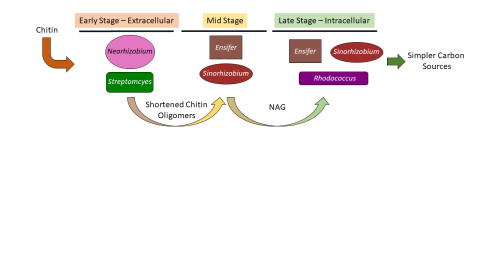
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. 16s data from MSC-2 growth. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33231 16s data from MSC-2 growth 3 fastq of 16s amplicon data of MSC2 1 csv file of raw...
Category
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Metatranscriptomic data from MSC-2. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33232 Metatranscriptomic data from MSC-2 12 fastq files (6 forward read, 6 reverse...
Category
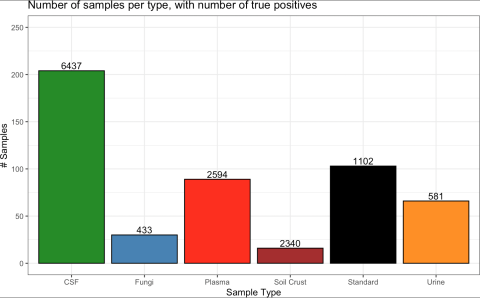
Data comprised of 204, 103, 89, 66, 30, and 16 gas chromatography mass spectrometry (GC-MS) runs from human cerebrospinal fluid (CSF), standards, human blood plasma, human urine, fungi species (A. niger, A. nidulans, T. reesei), and soil crust, respectively Across 4,523,251 candidate matches, 13,487...
Please cite as : Wu R., M.R. Davison, W.C. Nelson, M.L. Smith, M.S. Lipton, J.K. Jansson, and R.S. McClure, et al. 2023. HiC manuscript: bulk metagenomic data_MG_raw Upload. [Data Set] PNNL DataHub. https://doi.org/10.25584/1922087 The raw bulk metagenomic data were trimmed and de-novo assembled...