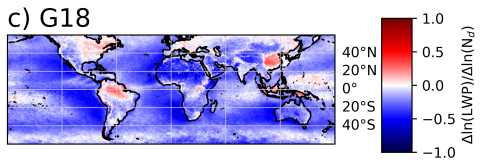
The SATELLITE_EAGLES_PNNL NetCDF dataset contains a suite of satellite- and reanalysis-derived atmospheric and surface parameters on a regular latitude–longitude grid for 6 different spatial resolutions (10, 5, 1, 0.5, 0.1, and 0.05 degree resolutions). The dataset includes core geophysical fields...
Filter results
Category
- Scientific Discovery (404)
- Biology (287)
- Earth System Science (167)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (14)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (28)
- Omics (26)
- Mass Spectrometry (25)
- Soil Microbiology (25)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- Synthetic (14)
- Genomics (13)
- sequencing (13)
- West Nile virus (13)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Machine Learning (8)
- Microbiome (8)
This data package contains processed LC-MS proteomics results and analysis scripts associated with the paper "Assessing Degenerate Peptide Resolution Methods using a Ground Truth Dataset". In this study, we designed an artificial microbial community to create a ground truth, simulated metaproteomic...
Category
Raw NMR data for the three trials measuring the distance between 15N in the backbone of glutamic acid in FD31* to 13C in the calcite surface using REDOR NMR. Data collected on a 300 MHz instrument using a 5 mm HCN probe.
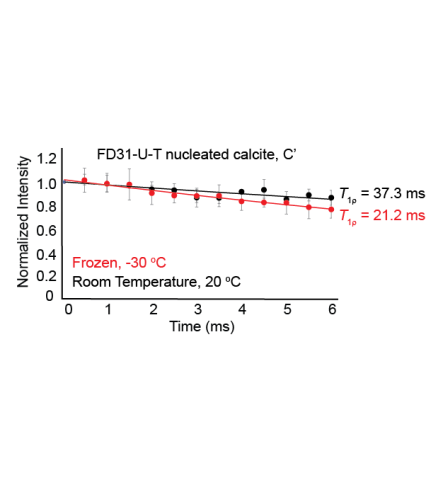
Raw data for all three trials used to calculate the T1rho value for FD31-U-T nucleated calcite at both room temperature and frozen. This data shows that the FD31 backbone carbonyl in threonine is immobile in nucleated calcite on the millisecond timescale at both temperatures.
Circular dichroism data showing that both FD31 and FD31* adopt identical helical secondary structures in solution.
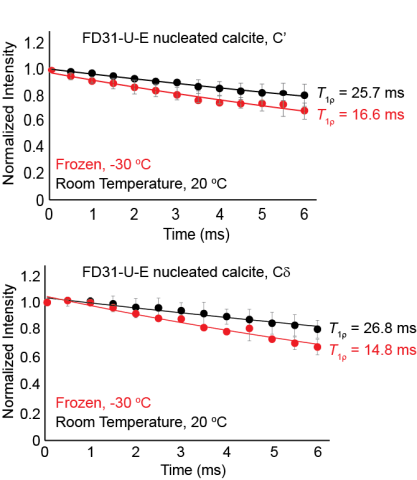
Raw data for all three trials used to calculate the T1rho value for FD31-U-E nucleated calcite at both room temperature and frozen. This data shows that both the FD31 backbone and sidechain carbonyls in glutamic are immobile in nucleated calcite on the millisecond timescale at both temperatures.
Solution NMR spectra of 5-13C-Glu amino acid used to synthesize FD31*-Edelta samples for solid state NMR. Spectra verifies that the starting material was only isotopically enriched at position 5.
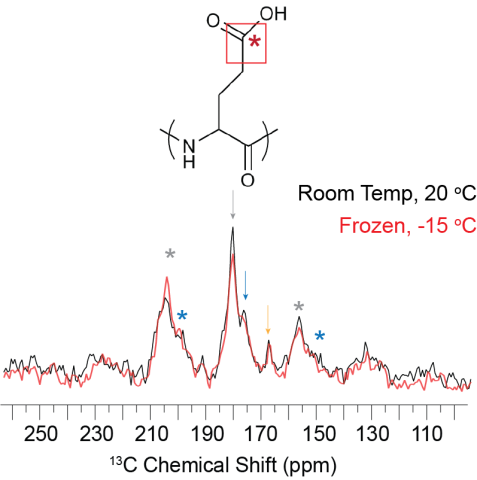
Room temperature and frozen CP-MAS spectra of FD31*-Edelta nucleated calcite showing the overall CSA pattern does not change as a function of temperature. The FD31* Glu sidechain carbonyl appears to be mobile on the ~500 microsecond timescale under both temperature conditions.
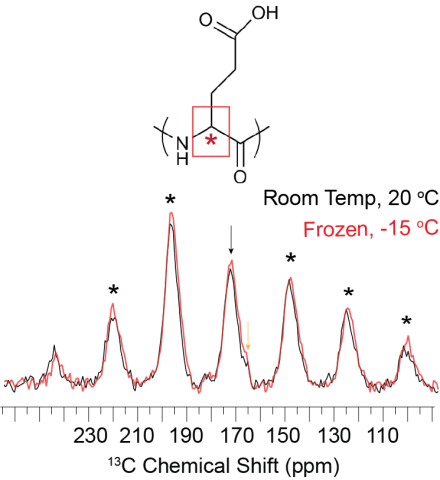
Room temperature and frozen CP-MAS spectra of FD31*-E' nucleated calcite showing the overall CSA pattern does not change as a function of temperature. The FD31* Glu backbone carbonyl appears to be immobile on the ~500 microsecond timescale under both temperature conditions.
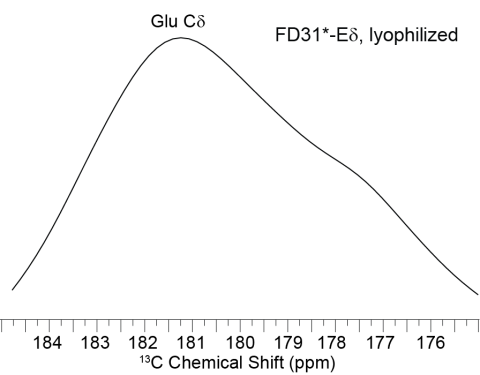
Solid state NMR CP-MAS 1H{13C} spectrum of FD31*-Edelta in the lyophilized state without calcite, showing that there are two distinct populations present even without surface.
The raw NMR data files for all three T1rho trials at room temperature for the FD31*-E' nucleated calcite sample showing almost no mobility for the Glu backbone carbonyls in FD31* on the millisecond timescale.
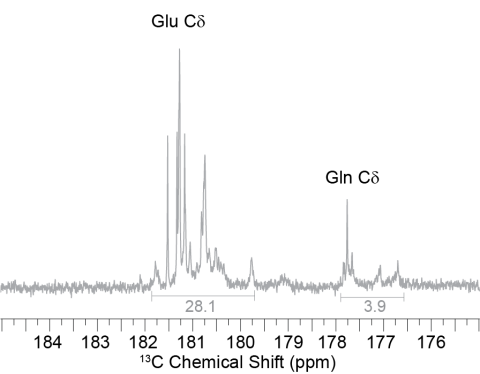
Solution NMR spectra of FD31*-Edelta dissolve in D2O, showing the distribution of peaks for glutamic acid (~180 ppm) and glutamine (~174 ppm), the latter of which was labeled via scrambling.
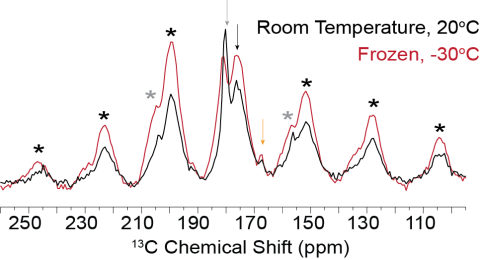
Room temperature and frozen CP-MAS spectra of FD31*-U-E nucleated calcite showing the overall CSA pattern does undergo slight perturbations as a function of temperature. While the overall CSA pattern does suggest the backbone carbonyl of Glu is immobile and the sidechain carbonyl of Glu is mobile on...
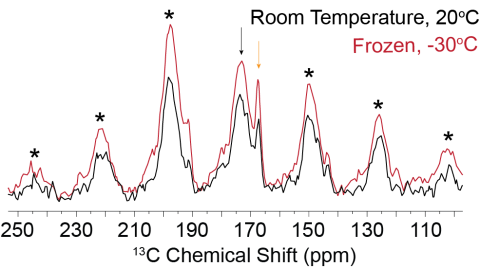
Comparison of frozen and room temperature 1H-13C CP-MAS spectra for FD31-U-T nucleated calcite show that the threonine backbone carbonyl is immobile at both temperatures on the ~500 microsecond timescale. A slight decrease in signal to noise between the room temperature and frozen spectra does...
The raw NMR data files for all three T1rho trials at room temperature for the FD31*-Edelta nucleated calcite sample showing almost no mobility for the Glu sidechain carbonyls in FD31* on the millisecond timescale.