Data Sample
Category
Description
Please cite as: Zegeye E., C.J. Brislawn, Y. Farris, S.J. Fansler, K.S. Hofmockel, J.K. Jansson, and A.T. Wright, et al. 2019. WA-IsoC_NAG.1.0 (Amplicon 16S/ITS, WA). [Data Set] PNNL DataHub. https://dx.doi.org/10.25584/data.2019-02.700/1506698
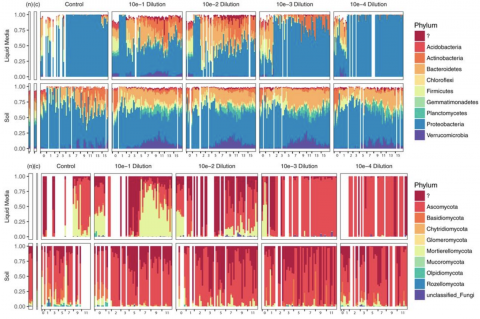
Investigation of the successional dynamics of a soil microbiome during 21-weeks of enrichment on chitin and its monomer, N-acetylglucosamine (NAG). Soil community succession was observed and analyzed over time in both a physically heterogeneous soil matrix as well as a homogeneous liquid medium. Soil sample collections were taken from Prosser, Washington, as a basis for these studies and this dataset represents amplicon (16S and ITS).
Amplicon sequencing of PCR products for investigating genetic variant identification and characterization of specific genomic regions. The versions described in this paper are the first version. Dataset package includes amplicon 16S and ITS sequencing files. To subselect from the folders listed below for download, please visit the Open Science Framework at the following link https://osf.io/6d5kz/. Supplementary data information is listed below under the data dictionary along with a download key for the items listed.
Items below available from Download Button:
IAREC, WA 16S Sequencing (fastq.gz)
- 180215 [2.47 GB; 659 items]
- 180317 [1.90 GB; 321 items]
- 180428 [1.69 GB; 321 items]
- 180529 [2.02 GB; 481 items]
IAREC, WA ITS Sequencing (fastq.gz)
- 180217 [1.14 GB; 659 items]
- 180423 [2.81 GB; 641 items]