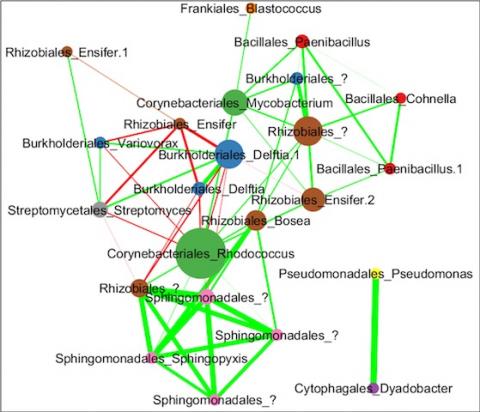
The soil microbiome is central to the cycling of carbon and other nutrients and to the promotion of plant growth. Despite its importance, analysis of the soil microbiome is difficult due to its sheer complexity, with thousands of interacting species. Here, we reduced this complexity by developing model soil microbial consortia that are simpler and more amenable to experimental analysis but still represent important microbial functions of the native soil ecosystem. Samples were collected from an arid grassland soil and microbial communities (consisting mainly of bacterial species) were enriched on agar plates containing chitin as the main carbon source. Chitin was chosen because it is an abundant carbon and nitrogen polymer in soil that often requires the coordinated action of several microorganisms for complete metabolic degradation. Several soil consortia were derived that had tractable richness (30–50 OTUs) with diverse phyla representative of the native soil, including Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia. The resulting consortia could be stored as glycerol or lyophilized stocks at −80°C and revived while retaining community composition, greatly increasing their use as tools for the research community at large. One of the consortia that was particularly stable was chosen as a model soil consortium (MSC-1) for further analysis. MSC-1 species interactions were studied using both pairwise co-cultivation in liquid media and during growth in soil under several perturbations. Co-abundance analyses highlighted interspecies interactions and helped to define keystone species, including Mycobacterium, Rhodococcus, and Rhizobiales taxa. These experiments demonstrate the success of an approach based on naturally enriching a community of interacting species that can be stored, revived, and shared. The knowledge gained from querying these communities and their interactions will enable better understanding of the soil microbiome and the roles these interactions play in this environment.
The versions described in this paper are the first version. Dataset package includes amplicon 16S rRNA fastq files, supplementary information, MIxS/MIMARKS.soil.5.0 metadata information, R markdown, and package “Read Me” file.
Projects (1)
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
45