Filter results
Category
- (-) Human Health (102)
- (-) Computational Research (23)
- (-) Visual Analytics (6)
- Scientific Discovery (307)
- Biology (198)
- Earth System Science (136)
- Integrative Omics (73)
- Microbiome Science (42)
- National Security (21)
- Computing & Analytics (14)
- Chemistry (10)
- Energy Resiliency (9)
- Data Analytics & Machine Learning (8)
- Materials Science (7)
- Chemical & Biological Signatures Science (5)
- Computational Mathematics & Statistics (5)
- Weapons of Mass Effect (5)
- Atmospheric Science (4)
- Coastal Science (4)
- Ecosystem Science (4)
- Renewable Energy (4)
- Data Analytics & Machine Learning (3)
- Plant Science (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Energy Storage (2)
- Grid Cybersecurity (2)
- Solar Energy (2)
- Bioenergy Technologies (1)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (30)
- Multi-Omics (28)
- Health (23)
- Virus (23)
- Viruses (23)
- MERS-CoV (18)
- Mus musculus (18)
- West Nile virus (13)
- Ebola (11)
- Influenza A (11)
- Resource Metadata (8)
- Microarray (7)
- Omics (7)
- Human Interferon (6)
- Omics-LHV Project (6)
- Type 1 Diabetes (6)
- Autoimmunity (5)
- Machine Learning (5)
- Biomarkers (4)
- Molecular Profiling (4)
- Processed Data (4)
- Proteomics (4)
- Software Data Analysis (4)
- Genomics (3)
Exhaled breath condensate proteomics represent a low-cost, non-invasive alternative for examining upper respiratory health. EBC has previously been used for the discovery and validation of detected exhaled volatiles and non-volatile biomarkers of disease related to upper respiratory system distress...
Dr. Gao obtained her Ph.D degree in Chemistry from institute of chemistry, Chinese Academy of Science. His Ph.D research focused on multiscale modeling of morphology and properties of polymeric materials, polymer processing and unveiling the process–properties relationships. (atomic to coarse...
Category
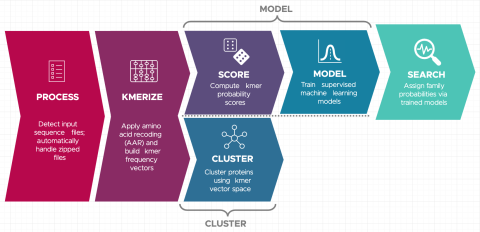
Christine H Chang, William C Nelson, Abby Jerger, Aaron T Wright, Robert G Egbert, Jason E McDermott, Snekmer: a scalable pipeline for protein sequence fingerprinting based on amino acid recoding, Bioinformatics Advances , Volume 3, Issue 1, 2023, vbad005, https://doi.org/10.1093/bioadv/vbad005...
Omics-LHV, West Nile Experiment WCD003 The purpose of this West Nile experiment was to obtain samples for omics analysis in mouse dendritic cell response to wild-type West Nile virus (WNV). Overall Design: Mouse dendritic cells (2 x 10^5) were treated with wild-type WNV and collected in parallel...
Category
Omics-LHV, West Nile Experiment WCN004 The purpose of this West Nile experiment was to obtain samples for omics analysis in mouse cerebral cortex neurons in response to wild-type West Nile Virus (WNV; WNV-NY99 382) and mutant WNV-E218A (WNV-NY99 382) viral infection. Overall Design: Mouse cortical...
Category
Omics-LHV, West Nile Experiment WGCN004 The purpose of this West Nile experiment was to obtain samples for omics analysis in primary mouse granule neuron cells infected with wild type West Nile virus (WNV-NY99 clone 382, WNVWT) and mutant virus (WNVE218A). Overall Design: Granule cell neurons from...
Category
The Sequel II System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by PacBio , and is designed for high throughput, production-scale sequencing laboratories. Originally released in 2015, the Sequel system provides Single Molecule, Real-Time (SMRT) sequencing core...
Human infections caused by viral pathogens trigger a complex gamut of host responses that limit disease, resolve infection, generate immunity, and contribute to severe disease or death. Here, we present experimental methods and multi-omics data capture approaches representing the global host...
Category
Category
The Human Islet Research Network (HIRN) is a large consortia with many research projects focused on understanding how beta cells are lost in type 1 diabetics (T1D) with a goal of finding how to protect against or replace the loss of functional beta cells. The consortia has multiple branches of...
Datasets
0
LIQUID Software Overview LIQUID provides users with the capability to process high throughput data and contains a customizable target library and scoring model per project needs. The graphical user interface provides visualization of multiple lines of spectral evidence for each lipid identification...
Category
pmartR Software Overview The pmartR package provides a single software tool for QC (filtering and normalization), exploratory data analysis (EDA), and statistical analysis (robust to missing data) and includes numerous visualization capabilities of mass spectrometry (MS) omics data (proteomic...
Fusarium sp. DS682 Proteogenomics Statistical Data Analysis of SFA dataset download: 10.25584/KSOmicsFspDS682/1766303 . GitHub Repository Source: https://github.com/lmbramer/Fusarium-sp.-DS-682-Proteogenomics MaxQuant Export Files (txt) Trelliscope Boxplots (jsonp) Fusarium Report (.Rmd, html)...
Last updated on 2023-02-23T19:37:46+00:00 by LN Anderson Snekmer: A scalable pipeline for protein sequence fingerprinting using amino acid recoding (AAR) Snekmer is a software package designed to reduce the representation of protein sequences by combining amino acid reduction (AAR) with the kmer...