Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL003 The purpose of this experiment was to evaluate the human host response to wild-type infectious clone of Middle Eastern Respiratory Syndrome coronavirus (icMERS-CoV) infection and mockulum. Sample data was obtained...
Filter results
Category
- (-) Human Health (70)
- Scientific Discovery (179)
- Biology (107)
- Earth System Science (89)
- Integrative Omics (52)
- Microbiome Science (16)
- National Security (14)
- Computing & Analytics (10)
- Energy Resiliency (7)
- Computational Research (6)
- Visual Analytics (6)
- Atmospheric Science (3)
- Coastal Science (3)
- Computational Mathematics & Statistics (3)
- Data Analytics & Machine Learning (3)
- Data Analytics & Machine Learning (3)
- Renewable Energy (3)
- Chemical & Biological Signatures Science (2)
- Cybersecurity (2)
- Distribution (2)
- Ecosystem Science (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Weapons of Mass Effect (2)
- Bioenergy Technologies (1)
- Chemistry (1)
- Computational Mathematics & Statistics (1)
- Energy Efficiency (1)
- Energy Storage (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Plant Science (1)
- Solar Energy (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Virology (64)
- Immune Response (48)
- Time Sampled Measurement Datasets (45)
- Gene expression profile data (42)
- Differential Expression Analysis (41)
- Homo sapiens (30)
- Mass spectrometry data (24)
- Multi-Omics (23)
- MERS-CoV (16)
- Mus musculus (15)
- Health (14)
- Virus (14)
- Viruses (14)
- West Nile virus (11)
- Ebola (9)
- Influenza A (9)
- Resource Metadata (8)
- Microarray (5)
- Human Interferon (4)
- Response to type I interferon (3)
- Epigenetics (2)
- Machine Learning (2)
- Processed Data (2)
- Response to type I & type II interferon (2)
- Type 1 Diabetes (2)
- Viral Experiment (2)
- EBC (1)
- Proteomics (1)
- Quantification (1)
- TMT (1)
Biomedical Resilience & Readiness in Adverse Operating Environments (BRAVE) Project: Exhaled Breath Condensate (EBC) TMT Proteomic Transformation Data Exhaled breath condensate (EBC) represents a low-cost and non-invasive means of examining respiratory health. EBC has been used to discover and...
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL105 Metadata The purpose of this experiment was to evaluate the host epigenetic response to Influenza A virus (subtype H5N1) infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL106 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to Influenza A virus (subtype H5N1) infection. Samples were obtained from human lung adenocarcinoma cell line (Calu...
Category
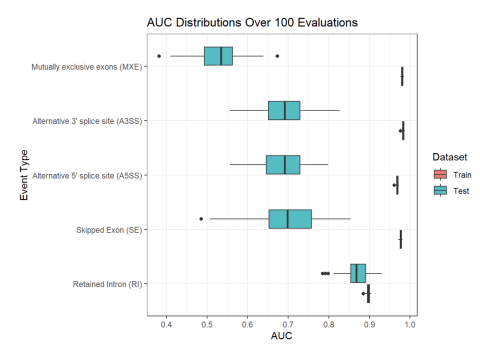
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...
Category
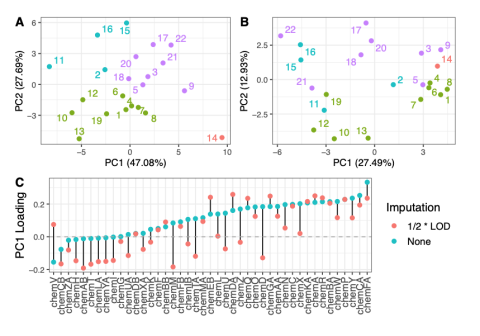
This data is supplementary to the manuscript Expanding the access of wearable silicone wristbands in community-engaged research through best practices in data analysis and integration by Lisa M. Bramer, Holly M. Dixon, David J. Degnan, Diana Rohlman, Julie B. Herbstman, Kim A. Anderson, and Katrina...
A total of 172 children from the DAISY study with multiple plasma samples collected over time, with up to 23 years of follow-up, were characterized via proteomics analysis. Of the children there were 40 controls and 132 cases. All 132 cases had measurements across time relative to IA. Sampling was...
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
Comprised of 6,426 sample runs, The Environmental Determinants of Diabetes in the Young (TEDDY) proteomics validation study constitutes one of the largest targeted proteomics studies in the literature to date. Making quality control (QC) and donor sample data available to researchers aligns with...