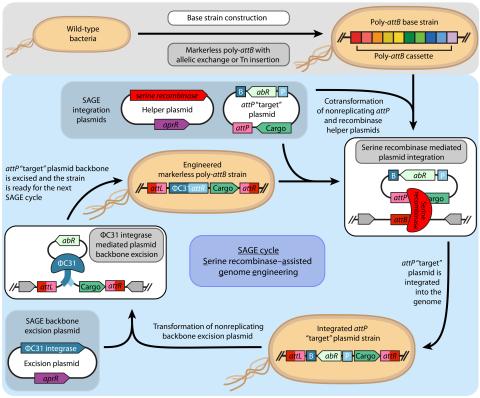
Elmore JR, Dexter GN, Baldino H, Huenemann JD, Francis R, Peabody GL 5th, Martinez-Baird J, Riley LA, Simmons T, Coleman-Derr D, Guss AM, Egbert RG. High-throughput genetic engineering of nonmodel and undomesticated bacteria via iterative site-specific genome integration. Sci Adv. 2023 Mar 10;9(10)...
Filter results
Content type
Tags
- (-) Statistical Expression Analysis (2)
- (-) Data Analysis (1)
- (-) DOE (1)
- Omics (9)
- PerCon SFA (9)
- High Throughput Sequencing (8)
- Genomics (7)
- Sequencer System (5)
- Synthetic Biology (5)
- Mass Spectrometry (3)
- A. pittii SO1 (2)
- Amplicon Sequencing (2)
- Biological and Environmental Research (2)
- Imaging (2)
- Long Read Sequencer (2)
- Mass spectrometry data (2)
- Proteomics (2)
- RNA Sequence Analysis (2)
- Sorghum bicolor (2)
- Spectroscopy (2)
- Whole Genome Sequencing (2)
- Environmental Microbiome Science (1)
- Genomic Sciences Program (1)
- Lipidomics (1)
- Machine Learning (1)
- Mass Spectrometer (1)
- metabolomics (1)
- Output Databases (1)
- Science Focus Area Project (1)
- Soil Microbiology (1)
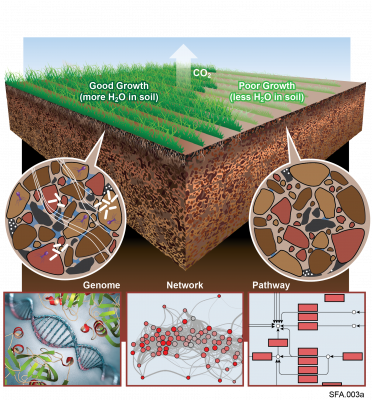
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
23
Fusarium sp. DS682 Proteogenomics Statistical Data Analysis of SFA dataset download: 10.25584/KSOmicsFspDS682/1766303 . GitHub Repository Source: https://github.com/lmbramer/Fusarium-sp.-DS-682-Proteogenomics MaxQuant Export Files (txt) Trelliscope Boxplots (jsonp) Fusarium Report (.Rmd, html)...
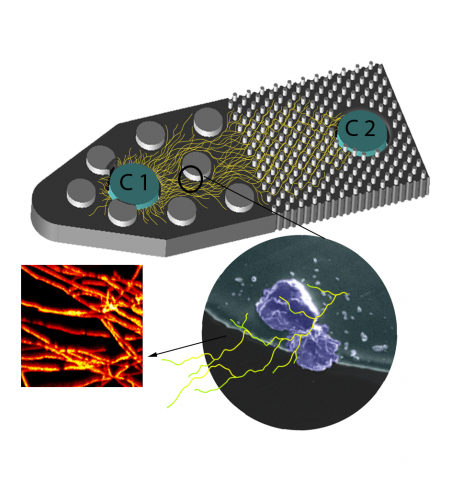
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project. Raw Measurement Data NCBI BioProject...