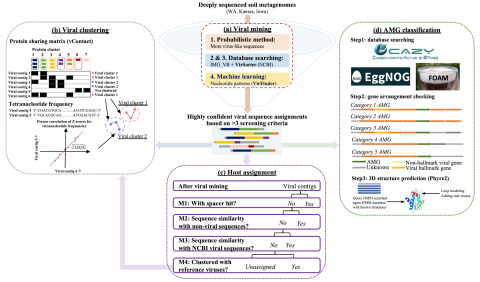
The following R source code was used for plotting figures of the viral communities detected from three grasslands soil metagenomes with a historical precipitation gradient ( WA-TmG.2.0 , KS-TmG.2.0 , IA-TmG.2.0 ) from project publication 'DNA viral diversity, abundance and functional potential vary...
Filter results
Category
- (-) Biology (38)
- Scientific Discovery (51)
- Earth System Science (23)
- Human Health (11)
- Microbiome Science (10)
- Materials Science (5)
- Computational Research (4)
- Atmospheric Science (1)
- Chemical & Biological Signatures Science (1)
- Chemistry (1)
- Computational Mathematics & Statistics (1)
- National Security (1)
- Plant Science (1)
- Weapons of Mass Effect (1)
Publication Type
Tags
- Viruses (11)
- Soil Microbiology (9)
- Health (8)
- Virology (8)
- Virus (8)
- PerCon SFA (5)
- Microbiome (4)
- Omics (4)
- sequencing (4)
- Fungi (3)
- Metagenomics (3)
- RNA Sequence Analysis (3)
- Synthetic Biology (3)
- Data Analysis (2)
- Genomics (2)
- High Throughput Sequencing (2)
- Mass Spectrometry (2)
- microbiome stability (2)
- Sorghum bicolor (2)
- Imaging (1)
- Lipidomics (1)
- metabolomics (1)
- metagenomics (1)
- Microarray (1)
- microbiome (1)
- Output Databases (1)
- Sequencing (1)
- soil microbiology (1)
- species volatility (1)
- Spectroscopy (1)
Fusarium sp. DS682 Proteogenomics Statistical Data Analysis of SFA dataset download: 10.25584/KSOmicsFspDS682/1766303 . GitHub Repository Source: https://github.com/lmbramer/Fusarium-sp.-DS-682-Proteogenomics MaxQuant Export Files (txt) Trelliscope Boxplots (jsonp) Fusarium Report (.Rmd, html)...
Soil fungi facilitate the translocation of inorganic nutrients from soil minerals to other microorganisms and plants. This ability is particularly advantageous in impoverished soils, because fungal mycelial networks can bridge otherwise spatially disconnected and inaccessible nutrient hotspots...
Category
Viral communities detected from three large grassland soil metagenomes with historically different precipitation moisture regimes.
Category
Code pertaining to the Soil Microbiome SFA Project publication data visualizations 'DNA viral diversity, abundance and functional potential vary across grassland soils with a range of historical moisture regimes' for processing publication data downloads.
Category
Viral communities detected from three large grassland soil metagenomes with historically different precipitation moisture regimes.
Category
Viral communities detected from three large grassland soil metagenomes with historically different precipitation moisture regimes.
Category
Exhaled breath condensate proteomics represent a low-cost, non-invasive alternative for examining upper respiratory health. EBC has previously been used for the discovery and validation of detected exhaled volatiles and non-volatile biomarkers of disease related to upper respiratory system distress...
The srpAnalytics modeling pipeline for the Superfund Research Program Analytics Portal.
Category
Citation: Gosline, S.J.C., Kim, D.N., Pande, P. et al. The Superfund Research Program Analytics Portal: linking environmental chemical exposure to biological phenotypes. Sci Data 10 , 151 (2023). https://doi.org/10.1038/s41597-023-02021-5 Funding Acknowledgments The research reported herein was...