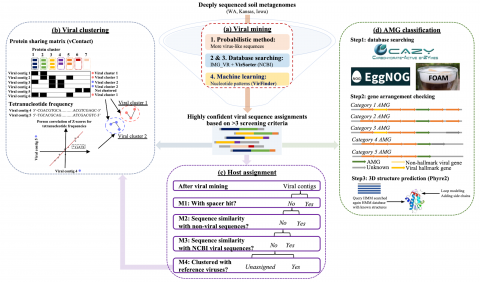
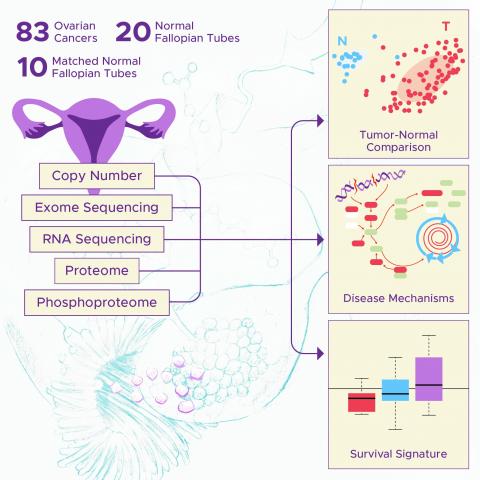
The srpAnalytics modeling pipeline for the Superfund Research Program Analytics Portal.
Filter results
Category
- (-) Biology (24)
- Scientific Discovery (35)
- Human Health (13)
- Earth System Science (11)
- Computational Research (10)
- Integrative Omics (8)
- National Security (5)
- Computing & Analytics (4)
- Data Analytics & Machine Learning (4)
- Microbiome Science (4)
- Energy Resiliency (2)
- Chemical & Biological Signatures Science (1)
- Chemistry (1)
- Coastal Science (1)
- Computational Mathematics & Statistics (1)
- Energy Efficiency (1)
- Energy Storage (1)
- Renewable Energy (1)
- Solar Energy (1)
- Weapons of Mass Effect (1)
Software Type
Tags
- Omics-LHV Project (6)
- Virology (6)
- Differential Expression Analysis (5)
- Gene expression profile data (5)
- Immune Response (5)
- Multi-Omics (5)
- Time Sampled Measurement Datasets (5)
- Autoimmunity (4)
- Biomarkers (4)
- Homo sapiens (4)
- Mass Spectrometry (4)
- Mass spectrometry data (4)
- Molecular Profiling (4)
- Omics (4)
- Type 1 Diabetes (4)
- Machine Learning (3)
- Mass spectrometry-based Omics (3)
- Mus musculus (3)
- Biological and Environmental Research (2)
- Ebola (2)
- Human Interferon (2)
- Influenza A (2)
- MERS-CoV (2)
- Predictive Modeling (2)
- Software Data Analysis (2)
- West Nile virus (2)
- Genomics (1)
- Output Databases (1)
- Proteomics (1)
- Statistics (1)
Code pertaining to the Soil Microbiome SFA Project publication data visualizations 'DNA viral diversity, abundance and functional potential vary across grassland soils with a range of historical moisture regimes' for processing publication data downloads.
Category
The Human Islet Research Network (HIRN) is a large consortia with many research projects focused on understanding how beta cells are lost in type 1 diabetics (T1D) with a goal of finding how to protect against or replace the loss of functional beta cells. The consortia has multiple branches of...
Datasets
0
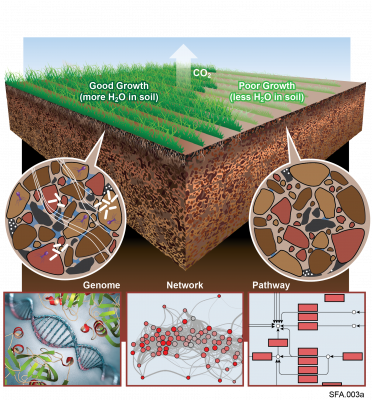
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
23
LIQUID Software Overview LIQUID provides users with the capability to process high throughput data and contains a customizable target library and scoring model per project needs. The graphical user interface provides visualization of multiple lines of spectral evidence for each lipid identification...
Category
pmartR Software Overview The pmartR package provides a single software tool for QC (filtering and normalization), exploratory data analysis (EDA), and statistical analysis (robust to missing data) and includes numerous visualization capabilities of mass spectrometry (MS) omics data (proteomic...
The following R source code was used for plotting figures of the viral communities detected from three grasslands soil metagenomes with a historical precipitation gradient ( WA-TmG.2.0 , KS-TmG.2.0 , IA-TmG.2.0 ) from project publication 'DNA viral diversity, abundance and functional potential vary...
Category
Fusarium sp. DS682 Proteogenomics Statistical Data Analysis of SFA dataset download: 10.25584/KSOmicsFspDS682/1766303 . GitHub Repository Source: https://github.com/lmbramer/Fusarium-sp.-DS-682-Proteogenomics MaxQuant Export Files (txt) Trelliscope Boxplots (jsonp) Fusarium Report (.Rmd, html)...
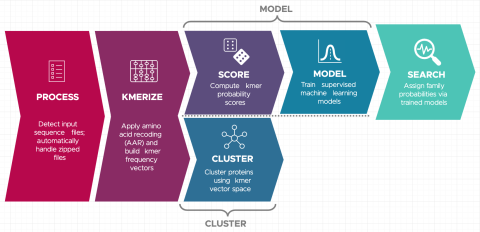
Last updated on 2023-02-23T19:37:46+00:00 by LN Anderson Snekmer: A scalable pipeline for protein sequence fingerprinting using amino acid recoding (AAR) Snekmer is a software package designed to reduce the representation of protein sequences by combining amino acid reduction (AAR) with the kmer...
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Omics-LHV Profiling of Host Interferon-Stimulated Response to Virus Infection Background The human host Interferon ( IFN ) alpha, beta, and gamma participate in the body's natural immune response to lethal virus infection and disease. The...
Category
Datasets
5
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Omics-LHV Profiling of Host Response to MERS-CoV Virus Infection Background Middle East Respiratory Syndrome coronavirus ( MERS-CoV ), part of the Coronaviridae family, is classified as a Category C priority pathogen by the National Institute...
Category
Datasets
15
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Omics-LHV Profiling of Host Response to Ebola Virus Infection Background Ebola virus ( EBOV ) is a high risk biological agent, belonging to the Flaviviridae family, and is classified as a Category A priority pathogen by the National Institute...
Category
Datasets
6
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Omics-LHV Profiling of Host Response to Influenza A Virus Infection Background Influenza A virus ( IAV ) is a high risk biological agent belonging to the Orthomyxoviridae family is classified as a Category C priority pathogen by the National...
Category
Datasets
8
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Omics-LHV Profiling of Host Response to West Nile Virus Infection Background West Nile virus ( WNV ) belongs to the mosquito-borne Flaviviridae family and is classified as a Category A priority pathogen by the National Institute of Allergy and...
Category
Datasets
11
Category
Datasets
1