Filter results
Software Type
Tags
- Omics (14)
- Genomics (5)
- High Throughput Sequencing (5)
- Mass Spectrometer (5)
- Sequencer System (5)
- Imaging (4)
- Mass Spectrometry (3)
- Microscopy (3)
- Proteomics (3)
- Lipidomics (2)
- Long Read Sequencer (2)
- Mass spectrometry data (2)
- metabolomics (2)
- Software Data Analysis (2)
- Data Analysis (1)
- DNA Sequence Analysis (1)
- EDX (1)
- Genome Analyzers (1)
- MALDI (1)
- Microarray (1)
- Microarray Platforms (1)
- Microbeam (1)
- Output Databases (1)
- SEM (1)
- Spectroscopy (1)
- Statistics (1)
- ToF-SIMS (1)
- X-Ray Diffraction (1)
- XANES (1)
- XRF (1)
The srpAnalytics modeling pipeline for the Superfund Research Program Analytics Portal.
Category
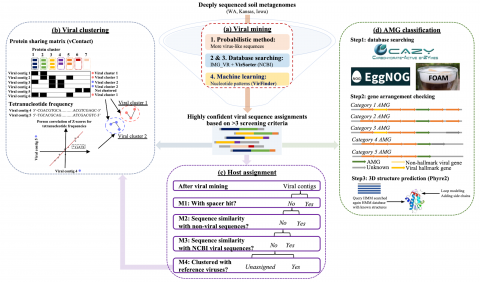
Code pertaining to the Soil Microbiome SFA Project publication data visualizations 'DNA viral diversity, abundance and functional potential vary across grassland soils with a range of historical moisture regimes' for processing publication data downloads.
Category
The Sequel II System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by PacBio , and is designed for high throughput, production-scale sequencing laboratories. Originally released in 2015, the Sequel system provides Single Molecule, Real-Time (SMRT) sequencing core...
LIQUID Software Overview LIQUID provides users with the capability to process high throughput data and contains a customizable target library and scoring model per project needs. The graphical user interface provides visualization of multiple lines of spectral evidence for each lipid identification...
Category
pmartR Software Overview The pmartR package provides a single software tool for QC (filtering and normalization), exploratory data analysis (EDA), and statistical analysis (robust to missing data) and includes numerous visualization capabilities of mass spectrometry (MS) omics data (proteomic...
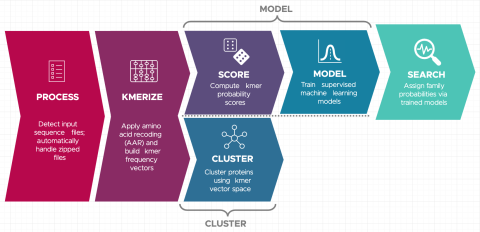
Last updated on 2023-02-23T19:37:46+00:00 by LN Anderson Snekmer: A scalable pipeline for protein sequence fingerprinting using amino acid recoding (AAR) Snekmer is a software package designed to reduce the representation of protein sequences by combining amino acid reduction (AAR) with the kmer...
The following R source code was used for plotting figures of the viral communities detected from three grasslands soil metagenomes with a historical precipitation gradient ( WA-TmG.2.0 , KS-TmG.2.0 , IA-TmG.2.0 ) from project publication 'DNA viral diversity, abundance and functional potential vary...
Category
Fusarium sp. DS682 Proteogenomics Statistical Data Analysis of SFA dataset download: 10.25584/KSOmicsFspDS682/1766303 . GitHub Repository Source: https://github.com/lmbramer/Fusarium-sp.-DS-682-Proteogenomics MaxQuant Export Files (txt) Trelliscope Boxplots (jsonp) Fusarium Report (.Rmd, html)...
The Illumina MiSeq System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for sequencing data acquisition using synthesis technology to provide an end-to-end solution (cluster generation, amplification, sequencing, and data analysis) in a...
Last updated on 2024-03-03T02:26:52+00:00 by LN Anderson The Thermo Scientific™ Q Exactive™ Plus Mass Spectrometer benchtop LC-MS/MS system combines quadruple precursor ion selection with high-resolution, accurate-mass (HRAM) Orbitrap detection to deliver exceptional performance and versatility...
Stanford Synchrotron Radiation Lightsource Experimental Station 14-3b is a bending magnet side station dedicated to X-Ray Imaging and Micro X-Ray Absorption Spectroscopy of biological, biomedical, materials, and geological samples. Station 14-3b is equipped with specialized instrumentation for XRF...
Category
The innovative Elstar™ electron column forms the basis of the Helios NanoLab’s outstanding high resolution imaging performance. The Elstar features unique technologies, such as constant power lenses for higher thermal stability, electrostatic scanning for faster, higher accurate imaging, and unique...
Category
The IONTOF TOF.SIMS 5 data source is a time-of-flight secondary ion mass spectrometer and powerful surface analysis tool used to investigate scientific questions in biological, environmental, and energy research. Among the most sensitive of surface analysis tools, it uses a high-vacuum technique...
The srpAnalytics modeling pipeline for processing new data for the OSU-PNNL Superfund Research Program Analytics Portal located at https://github.com/sgosline/srpAnalytics Data License MIT License Copyright (c) 2020 Sara JC Gosline . Permission is hereby granted, free of charge, to any person...