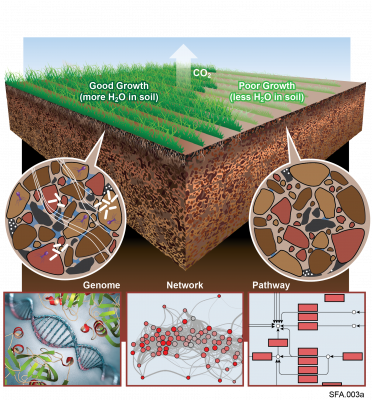
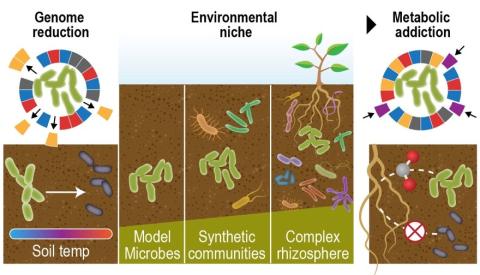
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Omics-LHV Profiling of Host Response to West Nile Virus Infection Background West Nile virus ( WNV ) belongs to the mosquito-borne Flaviviridae family and is classified as a Category A priority pathogen by the National Institute of Allergy and...
Category
Datasets
11