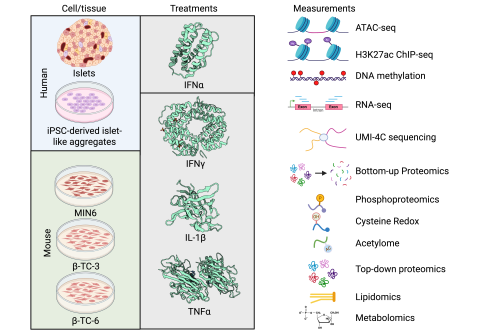
This data package consists of data from human islets (n=8) treated with IFNγ + IL-1β for 24 h and submitted to phosphoproteomics ( Table 1 ). Data contributor: Wei-Jun Qian: Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA, 99354, USA. Data repository: PXD011857...
Filter results
Category
- (-) Integrative Omics (96)
- (-) Materials Science (11)
- Scientific Discovery (402)
- Biology (285)
- Earth System Science (167)
- Human Health (113)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (14)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (55)
- Immune Response (51)
- Time Sampled Measurement Datasets (50)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (34)
- Multi-Omics (32)
- Mass spectrometry data (30)
- Mus musculus (19)
- MERS-CoV (18)
- West Nile virus (13)
- Influenza A (11)
- Omics (10)
- Ebola (9)
- Mass Spectrometry (8)
- Resource Metadata (7)
- Human Interferon (6)
- Microarray (6)
- Omics-LHV Project (6)
- Proteomics (6)
- Imaging (4)
- Mass Spectrometer (4)
- Microscopy (4)
- metabolomics (3)
- Response to type I interferon (3)
- Epigenetics (2)
- Lipidomics (2)
- Polymer Materials (2)
- Spectroscopy (2)
- Viral Experiment (2)
The data package consists of RNA-seq data from human islet-derived exosomes post IFNγ + IL-1β treatment of human islets (n=5) for 24 h ( Table 1 ). Data contributors: Preethi Krishnan& Carmella Evans-Molina: Department of Diabetes Immunology, Beckman Research Institute, City of Hope, Duarte, CA, USA...
Category
This data package consists of data from parallel experiments compared to Pck001 but performed with EndoC-βH1 cells instead of human islets. Cells were treated with IFNγ + IL-1β for 48 h and submitted to RNA-seq (n=5), ATAC-seq (n=5), DNA methylation (n=5), and H3K27Ac ChIP-seq (n=4). In addition...
Category
This data package consists of data from human islets (n=6) treated with IFNγ + IL-1β for 24 h and submitted to top-down proteomics ( Table 1 ). Data contributors: Ashley Ives, Wei-Jun Qian & James Fulcher: Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA, 99354, USA...
Category
This data package consists of data from sets of β-TC-3 cells (n=3) treated with IL-1β + IFNγ + TNFα for 24 h with or without ML351 and submitted to RNA-seq ( Table 1 ). ML351 is an inhibitor of 12-lipoxygenase, an enzyme that produces 12-HETE. Therefore, this treatment was done to study whether...
Category
This data package consists of data from β-TC-6 cells treated with IFNγ + IL-1β for 4, 8 or 24 h (n=4) and submitted to enrichment of cysteine-containing peptides (redox), phosphopeptides and acetylated peptides. This enabled the analysis of proteomics, redox cysteine, phosphoproteomics and...
Category
This data package contains processed LC-MS proteomics results and analysis scripts associated with the paper "Assessing Degenerate Peptide Resolution Methods using a Ground Truth Dataset". In this study, we designed an artificial microbial community to create a ground truth, simulated metaproteomic...
Category
Category
Category
Category
Category
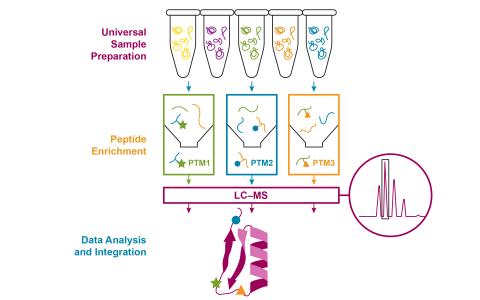
Created 2025-06-03T23:16:59+00:00 by LN Anderson .Currently pending DOI assignment for public release. Integrative SP3 Workflow for Multi-PTM Proteomics Profiling (PPI-TZ-DP0) The purpose of this experiment was to evaluate a new standardized method approach for proteomics sample processing designed...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
Omics-LHV, West Nile Experiment WGCN004 The purpose of this West Nile experiment was to obtain samples for omics analysis in primary mouse granule neuron cells infected with wild type West Nile virus (WNV-NY99 clone 382, WNVWT) and mutant virus (WNVE218A). Overall Design: Granule cell neurons from...