Omics-LHV, West Nile Experiment WCN004 The purpose of this West Nile experiment was to obtain samples for omics analysis in mouse cerebral cortex neurons in response to wild-type West Nile Virus (WNV; WNV-NY99 382) and mutant WNV-E218A (WNV-NY99 382) viral infection. Overall Design: Mouse cortical...
Filter results
Category
- (-) Scientific Discovery (404)
- (-) Weapons of Mass Effect (12)
- Biology (287)
- Earth System Science (167)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (14)
- Chemical & Biological Signatures Science (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (27)
- Omics (26)
- Mass Spectrometry (25)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Microbiome (8)
- Post-Translational Modifications (8)
- Microarray (7)
Omics-LHV, West Nile Experiment WCD003 The purpose of this West Nile experiment was to obtain samples for omics analysis in mouse dendritic cell response to wild-type West Nile virus (WNV). Overall Design: Mouse dendritic cells (2 x 10^5) were treated with wild-type WNV and collected in parallel...
Category
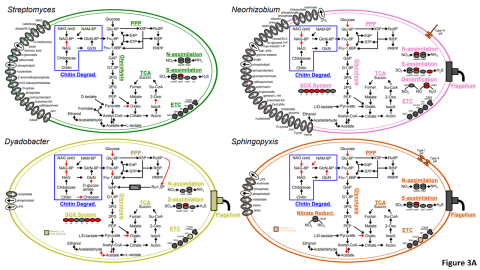
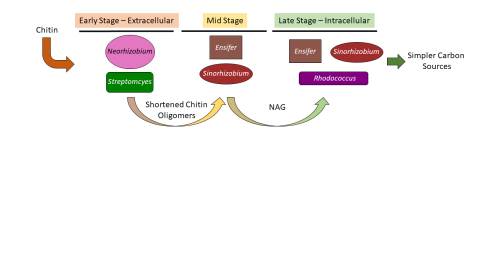
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes. [Data Set] PNNL DataHub. https://doi.org/10.25584/PNNLDH/1986536 Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes...
Category
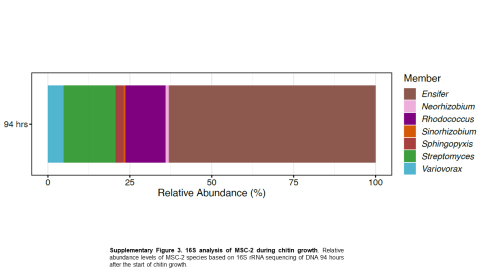
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. 16s data from MSC-2 growth. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33231 16s data from MSC-2 growth 3 fastq of 16s amplicon data of MSC2 1 csv file of raw...
Category
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Metatranscriptomic data from MSC-2. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33232 Metatranscriptomic data from MSC-2 12 fastq files (6 forward read, 6 reverse...
Category
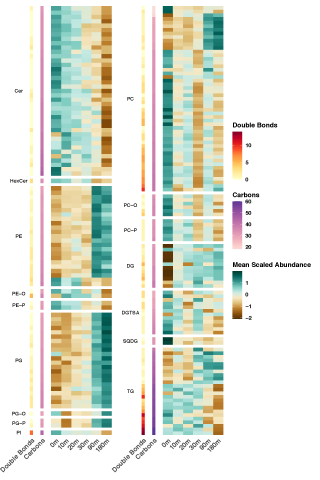
Rapid remodeling of the soil lipidome in response to a drying-rewetting event - Multi-Omics Data Package DOI Data package contents reported here are the first version and contain pre- and post-processed data acquisition and subsequent downstream analysis files using various data source instrument...
Category
ProxyTSPRD profiles are collected using NVIDIA Nsight Systems version 2020.3.2.6-87e152c and capture computational patterns from training deep learning-based time-series proxy-applications on four different levels: models (Long short-term Memory and Convolutional Neural Network), DL frameworks...
This data set provides the 16S microbial community composition via DNA sequence analysis from ingrowth peat and sand cores at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the...
Category
This data set provides the ITS fungal community composition via DNA and cDNA sequence analysis for peat and sand ingrowth cores collected before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE)...
Category
This data set provides ITS fungal community composition via DNA and cDNA sequence analysis at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were extracted using a Qiagen...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. KS-TmG.2.0 (Metagenome, KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/KSTmG2/1770332 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. WA-TmG.2.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG2/1770324 Soil samples were collected in triplicate in the fall of 2017 across the three grassland...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. IA-TmG.2.0 (Metagenome, IA). [Data Set] PNNL DataHub. https://doi.org/10.25584/IATmG2/1770333 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Last updated on 2025-01-16T22:12:33+00:00 by LN Anderson Fungal Monoisolate Multi-Omics Data Package DOI "KS4A-Omics1.0_FspDS68" Molecular mechanisms underlying fungal mineral weathering and nutrient translocation in low nutrient environments remain poorly resolved, due to the lack of a platform for...