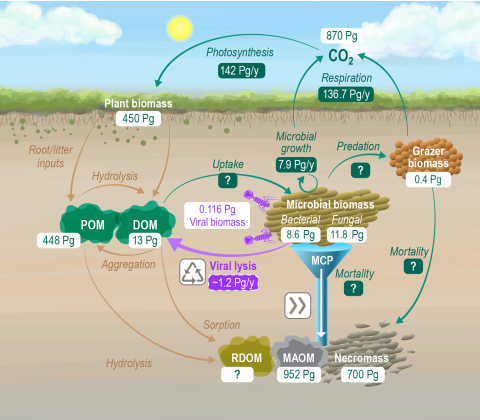
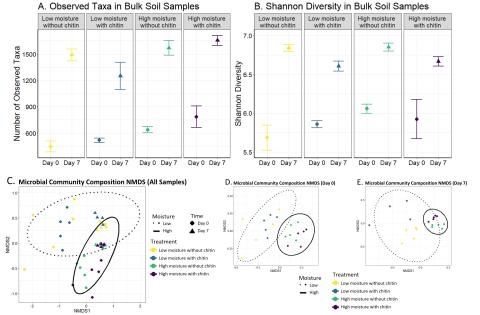
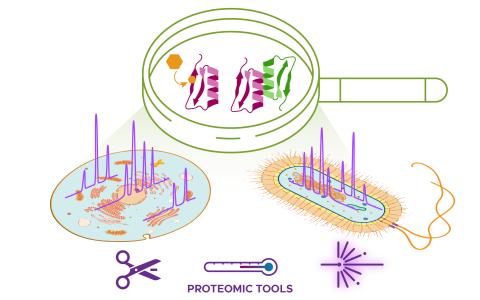
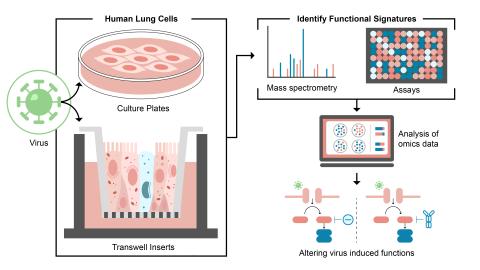
Please cite as : Rodríguez-Ramos, Josué, Natalie Sadler, Elias K. Zegeye, Yuliya Farris, Samuel Purvine, Sneha Couvillion, William C. Nelson, and Kirsten Hofmockel. Environmental matrix and moisture are key determinants of microbial phenotypes expressed in a reduced complexity soil-analog. 2024....
Filter results
Category
- Scientific Discovery (216)
- Biology (138)
- Earth System Science (107)
- Human Health (74)
- Integrative Omics (52)
- Microbiome Science (22)
- National Security (22)
- Computing & Analytics (12)
- Energy Resiliency (9)
- Chemical & Biological Signatures Science (8)
- Weapons of Mass Effect (8)
- Computational Research (6)
- Visual Analytics (6)
- Atmospheric Science (5)
- Data Analytics & Machine Learning (5)
- Materials Science (4)
- Renewable Energy (4)
- Coastal Science (3)
- Computational Mathematics & Statistics (3)
- Data Analytics & Machine Learning (3)
- Ecosystem Science (3)
- Bioenergy Technologies (2)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Storage (2)
- Grid Cybersecurity (2)
- Plant Science (2)
- Solar Energy (2)
- Subsurface Science (2)
- Transportation (2)
- Chemistry (1)
- Computational Mathematics & Statistics (1)
- Energy Efficiency (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Wind Energy (1)
Dataset Type
- Omics (107)
- Transcriptomics (53)
- Proteomics (51)
- Sequencing (41)
- Metabolomics (31)
- Lipidomics (28)
- Simulation Results (22)
- Amplicon (16S, ITS) (16)
- Metagenomics (9)
- Microarray (9)
- Computed Analysis (8)
- Microscopy (6)
- Genomics (4)
- Electron Microscopy (3)
- Geospacial (3)
- ChIP-seq (2)
- Mass Spectra (2)
- Phenomics (2)
- Staff (2)
- Cybersecurity (1)
- Electromagnetic Spectrum (1)
- Raw Image (1)
- Whole genome sequencing (WGS) (1)
Tags
- Virology (61)
- Immune Response (46)
- Time Sampled Measurement Datasets (46)
- Differential Expression Analysis (41)
- Gene expression profile data (40)
- Homo sapiens (37)
- Mass spectrometry data (25)
- Multi-Omics (24)
- Predictive Phenomics (17)
- MERS-CoV (16)
- Mus musculus (16)
- Soil Microbiology (15)
- Viruses (14)
- Health (13)
- Virus (13)
- West Nile virus (11)
- Mass Spectrometry (10)
- Proteomics (10)
- Synthetic (10)
- Influenza A (9)
- sequencing (9)
- TA1 (8)
- Ebola (7)
- Metagenomics (7)
- S. elongatus PCC 7942 (7)
- TA2 (7)
- Resource Metadata (6)
- Tandem Mass Tag (6)
- Global Analysis (5)
- Microarray (5)
Original data published and available from Zenodo at: https://doi.org/10.5281/zenodo.13839485 Please cite as : Zimmerman, A., & Hofmockel, K. (2024). Global terrestrial carbon pools and fluxes [Data set]. In Global Change Biology. Zenodo. https://doi.org/10.5281/zenodo.13839485 This file contains a...
Category
Please cite as : Schmidt A.J. 2023. MHTLS Cross-flow Heat Exchanger Temperature Performance Data. [Data Set] PNNL DataHub. 10.25584/2476481 The presentation and supporting information provide data on the performance of an engineering-scale cross-flow heat exchanger used in hydrothermal liquefaction...
Category
Please cite as : Burns C., and A.J. Schmidt. 2024. Rheology Investigations with Sludges from Metro Vancouver (Annacis Island Plant) Revision 1 . [Data Set] PNNL DataHub. 10.25584/2476482 Rheological investigation were performed with primary and secondary waste water treatment sludge. Using a...
Please cite as : Nicholas J Reichart, Sheryl Bell, Vanessa A. Garayburu-Caruso, Natalie Sadler, Sharon Zhao, Kirsten Hofmockel. Chitin Decomposition 16S rRNA gene fastq. 2024 . [Data Set] PNNL DataHub. 10.25584/2475041. This data is published under a CC0 license . The authors encourage data reuse...
Category
Currently pending public release.
Currently pending public release.
Created on 2024-10-16T17:27:40+00:00 by LN Anderson ; Last updated 2025-08-01T15:13:46+00:00. S. elongatus PCC 7942 LiP and TPP Structural Proteomics (JM-PB-DP3) The purpose of this experiment was to investigate structural alterations in proteins involved in central carbon metabolism and...
Currently pending public release.
Human Liver Epithelium Response to HCoV-229E Infection Epigenomics (ACS-DP4) The purpose of this experiment was to evaluate how wild-type Human coronavirus strain 229E (HCoV-229E) infection alters chromatin accessibility in infected cells. Sample data was obtained from mock-infected cells, UV...
Category
Human Primary Airway Epithelium Response to HCoV-229E Infection Transcriptomics (ACS-DP3) The purpose of this experiment was to evaluate the human host cellular response to wild-type Human coronavirus strain 229E (HCoV-229E) infection. Sample data was obtained for mock and infected (MOI 3) primary...
Category
Currently pending public release.
Category
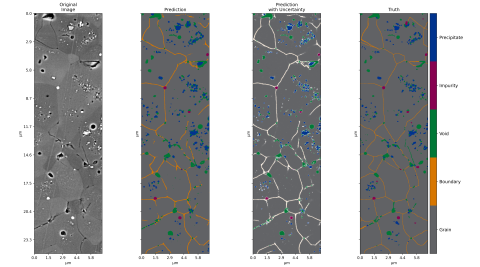
Irradiated/Irradiated_Calibrate/Iradiated_FL_TOPO_Meta • Performance metrics for all irradiated models in file starting with “all_notebooks” • Defect information within each model folder in csv files ending in “defects” and “defects_truth”, with “truth” for predicted images • Ripley results within...
Last updated by LN Anderson on 2025-09-03T17:50:50+00:00 S. elongatus PCC 7942 Carbon Metabolism Proteomics (MC-DP2) The purpose of this experiment was to understand the regulatory dynamics of carbon fixation in S. elongatus PCC 7942 under light and dark treatment conditions using a tandem mass tag...
Category
Currently pending public release.