"Deconstructing the Soil Microbiome into Reduced-Complexity Functional Modules"
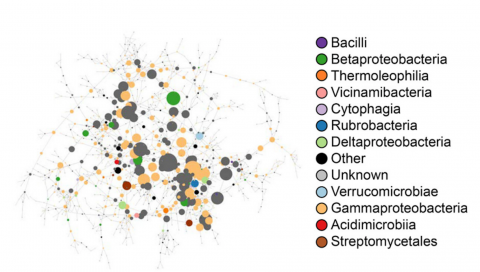
The soil microbiome represents one of the most complex microbial communities on the planet, encompassing thousands of taxa and metabolic pathways, rendering holistic analyses computationally intensive and difficult. Here, we developed an alternative approach in which the complex soil microbiome was broken into components (“functional modules”), based on metabolic capacities, for individual characterization. We hypothesized that reproducible, low-complexity communities that represent functional modules could be obtained through targeted enrichments and that, in combination, they would encompass a large extent of the soil microbiome diversity. Enrichments were performed on a starting soil inoculum with defined media based on specific carbon substrates, antibiotics, alternative electron acceptors under anaerobic conditions, or alternative growing conditions reflective of common field stresses. The resultant communities were evaluated through 16S rRNA amplicon sequencing. Less permissive modules (anaerobic conditions, complex polysaccharides, and certain stresses) resulted in more distinct community profiles with higher richness and more variability between replicates, whereas modules with simple substrates were dominated by fewer species and were more reproducible. Collectively, approximately 27% of unique taxa present in the liquid soil extract control were found across functional modules. Taxa that were underrepresented or undetected in the source soil were also enriched across the modules. Metatranscriptomic analyses were carried out on a subset of the modules to investigate differences in functional gene expression. These results demonstrate that by dissecting the soil microbiome into discrete components it is possible to obtain a more comprehensive view of the soil microbiome and its biochemical potential than would be possible using more holistic analyses. Code used to generate statistical inferences within the manuscript can be found under "Resources" listed below.
Linked Project Data
- Soil Metagenome Assembly: GCA_011392945.1
- RNA-Seq (metagenome): PRJNA597444 | BioSample: SAMN13675441 | WGS: WUUC00000000
- 16S Amplicon: PRJNA594403 | BioSample(s): SAMN13560171 - SAMN13560497 | SRA: SRP237537 (327 samples)
- Gene Expression Omnibus (70) and are available through GEO Series accession number GSE143587 (70 samples)
- Statistical inferences within the manuscript are publicly available at https://github.com/dtnaylor124/FunctionalModules
KEYWORDS: Functional modules, microbiome, reduced complexity, soil metatranscriptome, soil microbiome
Naylor D, Fansler S, Brislawn C, Nelson WC, Hofmockel KS, Jansson JK, McClure R. Deconstructing the Soil Microbiome into Reduced-Complexity Functional Modules. mBio. 2020 Jul 7;11(4):e01349-20.10.1128/mBio.01349-20
Projects (1)
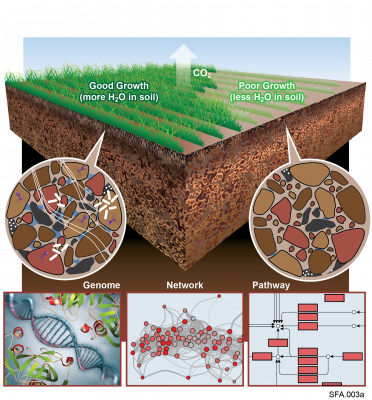
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
45